The architecture of a eukaryotic replisome
- PMID: 26524492
- PMCID: PMC4849863
- DOI: 10.1038/nsmb.3113
The architecture of a eukaryotic replisome
Abstract
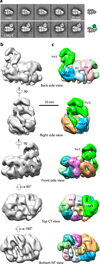
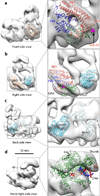
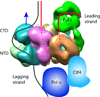
At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase is positioned in front to unwind DNA and that DNA polymerases trail behind the helicase. Here we used single-particle EM to directly image a Saccharomyces cerevisiae replisome. Contrary to expectations, the leading strand Pol ɛ is positioned ahead of CMG helicase, whereas Ctf4 and the lagging-strand polymerase (Pol) α-primase are behind the helicase. This unexpected architecture indicates that the leading-strand DNA travels a long distance before reaching Pol ɛ, first threading through the Mcm2-7 ring and then making a U-turn at the bottom and reaching Pol ɛ at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture and provides a basis for further structural and biochemical replisome studies.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- AG29979/AG/NIA NIH HHS/United States
- R37 GM038839/GM/NIGMS NIH HHS/United States
- GM109824/GM/NIGMS NIH HHS/United States
- R01 GM074985/GM/NIGMS NIH HHS/United States
- R01 GM111742/GM/NIGMS NIH HHS/United States
- R01 GM038839/GM/NIGMS NIH HHS/United States
- R35 GM131754/GM/NIGMS NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
- R01 AG029979/AG/NIA NIH HHS/United States
- P41 GM103314/GM/NIGMS NIH HHS/United States
- S10 RR025415/RR/NCRR NIH HHS/United States
- GM74985/GM/NIGMS NIH HHS/United States
- R01 GM115809/GM/NIGMS NIH HHS/United States
- GM103314/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM38839/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

