Autophagy mediates degradation of nuclear lamina
- PMID: 26524528
- PMCID: PMC4824414
- DOI: 10.1038/nature15548
Autophagy mediates degradation of nuclear lamina
Abstract
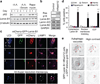
Macroautophagy (hereafter referred to as autophagy) is a catabolic membrane trafficking process that degrades a variety of cellular constituents and is associated with human diseases. Although extensive studies have focused on autophagic turnover of cytoplasmic materials, little is known about the role of autophagy in degrading nuclear components. Here we report that the autophagy machinery mediates degradation of nuclear lamina components in mammals. The autophagy protein LC3/Atg8, which is involved in autophagy membrane trafficking and substrate delivery, is present in the nucleus and directly interacts with the nuclear lamina protein lamin B1, and binds to lamin-associated domains on chromatin. This LC3-lamin B1 interaction does not downregulate lamin B1 during starvation, but mediates its degradation upon oncogenic insults, such as by activated RAS. Lamin B1 degradation is achieved by nucleus-to-cytoplasm transport that delivers lamin B1 to the lysosome. Inhibiting autophagy or the LC3-lamin B1 interaction prevents activated RAS-induced lamin B1 loss and attenuates oncogene-induced senescence in primary human cells. Our study suggests that this new function of autophagy acts as a guarding mechanism protecting cells from tumorigenesis.
Figures




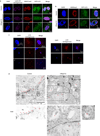

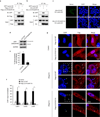

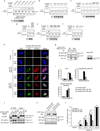
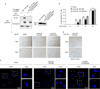

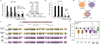


Comment in
-
Autophagy: Nuclear autophagy in tumour suppression.Nat Rev Mol Cell Biol. 2015 Dec;16(12):700-1. doi: 10.1038/nrm4091. Epub 2015 Nov 4. Nat Rev Mol Cell Biol. 2015. PMID: 26530389 No abstract available.
-
Autophagy Mediates Tumor Suppression via Cellular Senescence.Trends Cell Biol. 2016 Jan;26(1):1-3. doi: 10.1016/j.tcb.2015.11.001. Epub 2015 Nov 21. Trends Cell Biol. 2016. PMID: 26612212
-
Autophagy Devours the Nuclear Lamina to Thwart Oncogenic Stress.Dev Cell. 2015 Dec 7;35(5):529-530. doi: 10.1016/j.devcel.2015.11.016. Dev Cell. 2015. PMID: 26651287
References
Supplemental References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

