Incompatible Natures of the HIV-1 Envelope in Resistance to the CCR5 Antagonist Cenicriviroc and to Neutralizing Antibodies
- PMID: 26525792
- PMCID: PMC4704143
- DOI: 10.1128/AAC.02285-15
Incompatible Natures of the HIV-1 Envelope in Resistance to the CCR5 Antagonist Cenicriviroc and to Neutralizing Antibodies
Abstract
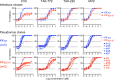
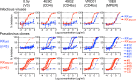
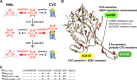
Cenicriviroc is a CCR5 antagonist which prevents human immunodeficiency virus type 1 (HIV-1) from cellular entry. The CCR5-binding regions of the HIV-1 envelope glycoprotein are important targets for neutralizing antibodies (NAbs), and mutations conferring cenicriviroc resistance may therefore affect sensitivity to NAbs. Here, we used the in vitro induction of HIV-1 variants resistant to cenicriviroc or NAbs to examine the relationship between resistance to cenicriviroc and resistance to NAbs. The cenicriviroc-resistant variant KK652-67 (strain KK passaged 67 times in the presence of increasing concentrations of cenicriviroc) was sensitive to neutralization by NAbs against the V3 loop, the CD4-induced (CD4i) region, and the CD4-binding site (CD4bs), whereas the wild-type (WT) parental HIV-1 strain KKWT from which cenicriviroc-resistant strain KK652-67 was obtained was resistant to these NAbs. The V3 region of KK652-67 was important for cenicriviroc resistance and critical to the high sensitivity of the V3, CD4i, and CD4bs epitopes to NAbs. Moreover, induction of variants resistant to anti-V3 NAb 0.5γ and anti-CD4i NAb 4E9C from cenicriviroc-resistant strain KK652-67 resulted in reversion to the cenicriviroc-sensitive phenotype comparable to that of the parental strain, KKWT. Resistance to 0.5γ and 4E9C was caused by the novel substitutions R315K, G324R, and E381K in the V3 and C3 regions near the substitutions conferring cenicriviroc resistance. Importantly, these amino acid changes in the CCR5-binding region were also responsible for reversion to the cenicriviroc-sensitive phenotype. These results suggest the presence of key amino acid residues where resistance to cenicriviroc is incompatible with resistance to NAbs. This implies that cenicriviroc and neutralizing antibodies may restrict the emergence of variants resistant to each other.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. 2013. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 341:1387–1390. doi:10.1126/science.1241475. - DOI - PMC - PubMed
-
- Schnur E, Noah E, Ayzenshtat I, Sargsyan H, Inui T, Ding FX, Arshava B, Sagi Y, Kessler N, Levy R, Scherf T, Naider F, Anglister J. 2011. The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide. J Mol Biol 410:778–797. doi:10.1016/j.jmb.2011.04.023. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

