Determining multiallelic complex copy number and sequence variation from high coverage exome sequencing data
- PMID: 26526070
- PMCID: PMC4630827
- DOI: 10.1186/s12864-015-2123-y
Determining multiallelic complex copy number and sequence variation from high coverage exome sequencing data
Abstract
Background: Copy number variation (CNV) is a major component of genomic variation, yet methods to accurately type genomic CNV lag behind methods that type single nucleotide variation. High-throughput sequencing can contribute to these methods by using sequence read depth, which takes the number of reads that map to a given part of the reference genome as a proxy for copy number of that region, and compares across samples. Furthermore, high-throughput sequencing also provides information on the sequence differences between copies within and between individuals.
Methods: In this study we use high-coverage phase 3 exome sequences of the 1000 Genomes project to infer diploid copy number of the beta-defensin genomic region, a well-studied CNV that carries several beta-defensin genes involved in the antimicrobial response, signalling, and fertility. We also use these data to call sequence variants, a particular challenge given the multicopy nature of the region.
Results: We confidently call copy number and sequence variation of the beta-defensin genes on 1285 samples from 26 global populations, validate copy number using Nanostring nCounter and triplex paralogue ratio test data. We use the copy number calls to verify the genomic extent of the CNV and validate sequence calls using analysis of cloned PCR products. We identify novel variation, mostly individually rare, predicted to alter amino-acid sequence in the beta-defensin genes. Such novel variants may alter antimicrobial properties or have off-target receptor interactions, and may contribute to individuality in immunological response and fertility.
Conclusions: Given that 81% of identified sequence variants were not previously in dbSNP, we show that sequence variation in multiallelic CNVs represent an unappreciated source of genomic diversity.
Figures
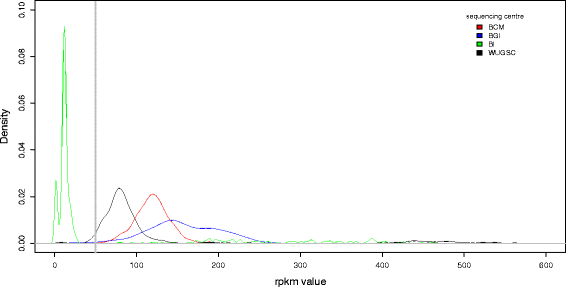
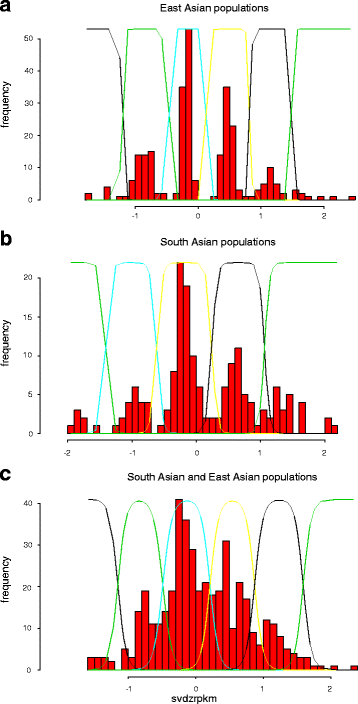
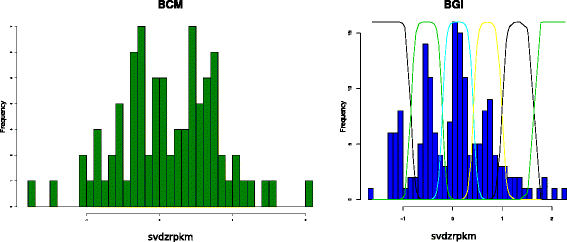
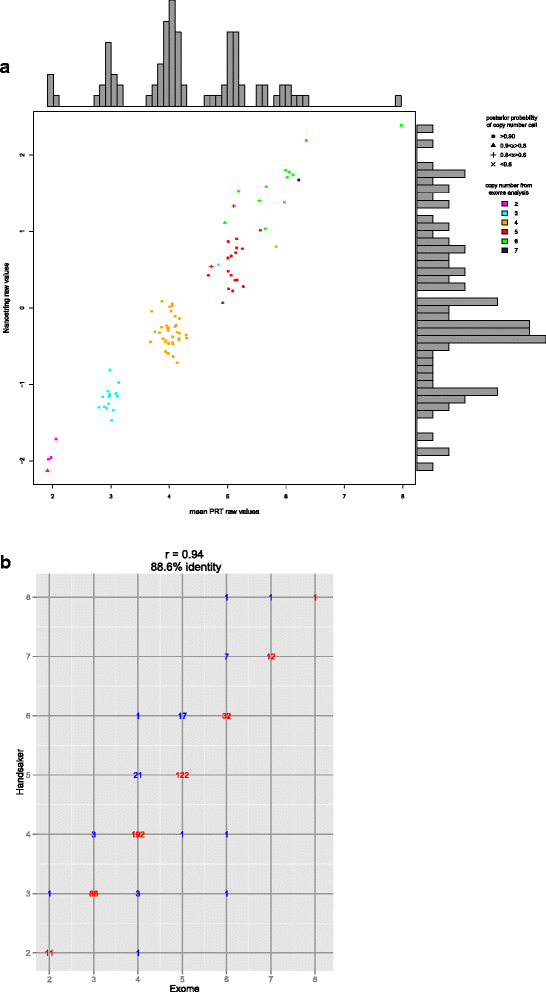

References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

