Cloning-free CRISPR
- PMID: 26527385
- PMCID: PMC4649464
- DOI: 10.1016/j.stemcr.2015.09.022
Cloning-free CRISPR
Abstract
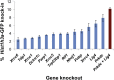
We present self-cloning CRISPR/Cas9 (scCRISPR), a technology that allows for CRISPR/Cas9-mediated genomic mutation and site-specific knockin transgene creation within several hours by circumventing the need to clone a site-specific single-guide RNA (sgRNA) or knockin homology construct for each target locus. We introduce a self-cleaving palindromic sgRNA plasmid and a short double-stranded DNA sequence encoding the desired locus-specific sgRNA into target cells, allowing them to produce a locus-specific sgRNA plasmid through homologous recombination. scCRISPR enables efficient generation of gene knockouts (∼88% mutation rate) at approximately one-sixth the cost of plasmid-based sgRNA construction with only 2 hr of preparation for each targeted site. Additionally, we demonstrate efficient site-specific knockin of GFP transgenes without any plasmid cloning or genome-integrated selection cassette in mouse and human embryonic stem cells (2%-4% knockin rate) through PCR-based addition of short homology arms. scCRISPR substantially lowers the bar on mouse and human transgenesis.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

