A 13 mer LNA-i-miR-221 Inhibitor Restores Drug Sensitivity in Melphalan-Refractory Multiple Myeloma Cells
- PMID: 26527748
- PMCID: PMC4775414
- DOI: 10.1158/1078-0432.CCR-15-0489
A 13 mer LNA-i-miR-221 Inhibitor Restores Drug Sensitivity in Melphalan-Refractory Multiple Myeloma Cells
Erratum in
-
Editor's Note: A 13 mer LNA-i-miR-221 Inhibitor Restores Drug Sensitivity in Melphalan-Refractory Multiple Myeloma Cells.Clin Cancer Res. 2024 May 15;30(10):2288. doi: 10.1158/1078-0432.CCR-24-1089. Clin Cancer Res. 2024. PMID: 38745477 No abstract available.
Abstract
Purpose: The onset of drug resistance is a major cause of treatment failure in multiple myeloma. Although increasing evidence is defining the role of miRNAs in mediating drug resistance, their potential activity as drug-sensitizing agents has not yet been investigated in multiple myeloma.
Experimental design: Here we studied the potential utility of miR-221/222 inhibition in sensitizing refractory multiple myeloma cells to melphalan.
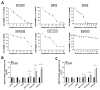
Results: miR-221/222 expression inversely correlated with melphalan sensitivity of multiple myeloma cells. Inhibition of miR-221/222 overcame melphalan resistance and triggered apoptosis of multiple myeloma cells in vitro, in the presence or absence of human bone marrow (BM) stromal cells. Decreased multiple myeloma cell growth induced by inhibition of miR-221/222 plus melphalan was associated with a marked upregulation of pro-apoptotic BBC3/PUMA protein, a miR-221/222 target, as well as with modulation of drug influx-efflux transporters SLC7A5/LAT1 and the ABC transporter ABCC1/MRP1. Finally, in vivo treatment of SCID/NOD mice bearing human melphalan-refractory multiple myeloma xenografts with systemic locked nucleic acid (LNA) inhibitors of miR-221 (LNA-i-miR-221) plus melphalan overcame drug resistance, evidenced by growth inhibition with significant antitumor effects together with modulation of PUMA and ABCC1 in tumors retrieved from treated mice.
Conclusions: Taken together, our findings provide the proof of concept that LNA-i-miR-221 can reverse melphalan resistance in preclinical models of multiple myeloma, providing the framework for clinical trials to overcome drug resistance, and improve patient outcome in multiple myeloma.
©2015 American Association for Cancer Research.
Conflict of interest statement
Figures

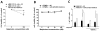



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

