In-Depth Assessment of Within-Individual and Inter-Individual Variation in the B Cell Receptor Repertoire
- PMID: 26528292
- PMCID: PMC4601265
- DOI: 10.3389/fimmu.2015.00531
In-Depth Assessment of Within-Individual and Inter-Individual Variation in the B Cell Receptor Repertoire
Abstract
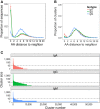
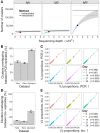
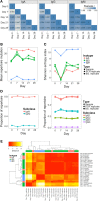
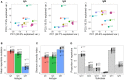
High-throughput sequencing of the B cell receptor (BCR) repertoire can provide rapid characterization of the B cell response in a wide variety of applications in health, after vaccination and in infectious, inflammatory and immune-driven disease, and is starting to yield clinical applications. However, the interpretation of repertoire data is compromised by a lack of studies to assess the intra and inter-individual variation in the BCR repertoire over time in healthy individuals. We applied a standardized isotype-specific BCR repertoire deep sequencing protocol to a single highly sampled participant, and then evaluated the method in 9 further participants to comprehensively describe such variation. We assessed total repertoire metrics of mutation, diversity, VJ gene usage and isotype subclass usage as well as tracking specific BCR sequence clusters. There was good assay reproducibility (both in PCR amplification and biological replicates), but we detected striking fluctuations in the repertoire over time that we hypothesize may be due to subclinical immune activation. Repertoire properties were unique for each individual, which could partly be explained by a decrease in IgG2 with age, and genetic differences at the immunoglobulin locus. There was a small repertoire of public clusters (0.5, 0.3, and 1.4% of total IgA, IgG, and IgM clusters, respectively), which was enriched for expanded clusters containing sequences with suspected specificity toward antigens that should have been historically encountered by all participants through prior immunization or infection. We thus provide baseline BCR repertoire information that can be used to inform future study design, and aid in interpretation of results from these studies. Furthermore, our results indicate that BCR repertoire studies could be used to track changes in the public repertoire in and between populations that might relate to population immunity against infectious diseases, and identify the characteristics of inflammatory and immunological diseases.
Keywords: B cell; B cell receptor repertoire; VDJ recombination; antibody diversity; genetic variation; immunoglobulin gene; immunoglobulin repertoire; reproducibility.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

