Population genomic datasets describing the post-vaccine evolutionary epidemiology of Streptococcus pneumoniae
- PMID: 26528397
- PMCID: PMC4622223
- DOI: 10.1038/sdata.2015.58
Population genomic datasets describing the post-vaccine evolutionary epidemiology of Streptococcus pneumoniae
Abstract
Streptococcus pneumoniae is common nasopharyngeal commensal bacterium and important human pathogen. Vaccines against a subset of pneumococcal antigenic diversity have reduced rates of disease, without changing the frequency of asymptomatic carriage, through altering the bacterial population structure. These changes can be studied in detail through using genome sequencing to characterise systematically-sampled collections of carried S. pneumoniae. This dataset consists of 616 annotated draft genomes of isolates collected from children during routine visits to primary care physicians in Massachusetts between 2001, shortly after the seven valent polysaccharide conjugate vaccine was introduced, and 2007. Also made available are a core genome alignment and phylogeny describing the overall population structure, clusters of orthologous protein sequences, software for inferring serotype from Illumina reads, and whole genome alignments for the analysis of closely-related sets of pneumococci. These data can be used to study both bacterial evolution and the epidemiology of a pathogen population under selection from vaccine-induced immunity.
Keywords: Bacterial genetics; Genetic variation; Molecular evolution; Respiratory tract diseases.
Conflict of interest statement
S.I.P. has investigator-initiated grants from Merck and Pfizer and has consulted for GlaxoSmithKline, Merck, Pfizer and Novartis. W.P.H. has consulted for GlaxoSmithKline. M.L. has consulted for Pfizer and Novartis.
Figures
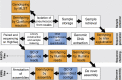

Dataset use reported in
- doi: 10.1542/peds.2008-3099
- doi: 10.1542/peds.112.4.862
- doi: 10.1542/peds.2004-2338
- doi: 10.1099/00221287-144-11-3049
- doi: 10.1086/510249
- doi: 10.1016/j.vaccine.2011.09.075
- doi: 10.1038/ng.2625
- doi: 10.1038/ncomms6471
References
Data Citations
-
- Croucher N.J. 2009. International Nucleotide Sequence Database . FM211187
-
- Croucher N.J. 2015. Dryad. http://dx.doi.org/10.5061/dryad.t55gq - DOI
-
- Croucher N.J. 2013. International Nucleotide Sequence Database . PRJEB2632
-
- Croucher N.J. 2015. Microreact. http://microreact.org/project/NJwviE7F
References
-
- Weintraub A. Immunology of bacterial polysaccharide antigens. Carbohydrate Research 338, 2539–2547 (2003). - PubMed
-
- Austrian R. Pneumococcal otitis media and pneumococcal vaccines, a historical perspective. Vaccine 19 (Suppl 1): S71–S77 (2000). - PubMed
-
- Whitney C. G. et al. Effectiveness of seven-valent pneumococcal conjugate vaccine against invasive pneumococcal disease: a matched case-control study. Lancet 368, 1495–1502 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

