Genome-Wide Association Studies of the Human Gut Microbiota
- PMID: 26528553
- PMCID: PMC4631601
- DOI: 10.1371/journal.pone.0140301
Genome-Wide Association Studies of the Human Gut Microbiota
Abstract
The bacterial composition of the human fecal microbiome is influenced by many lifestyle factors, notably diet. It is less clear, however, what role host genetics plays in dictating the composition of bacteria living in the gut. In this study, we examined the association of ~200K host genotypes with the relative abundance of fecal bacterial taxa in a founder population, the Hutterites, during two seasons (n = 91 summer, n = 93 winter, n = 57 individuals collected in both). These individuals live and eat communally, minimizing variation due to environmental exposures, including diet, which could potentially mask small genetic effects. Using a GWAS approach that takes into account the relatedness between subjects, we identified at least 8 bacterial taxa whose abundances were associated with single nucleotide polymorphisms in the host genome in each season (at genome-wide FDR of 20%). For example, we identified an association between a taxon known to affect obesity (genus Akkermansia) and a variant near PLD1, a gene previously associated with body mass index. Moreover, we replicate a previously reported association from a quantitative trait locus (QTL) mapping study of fecal microbiome abundance in mice (genus Lactococcus, rs3747113, P = 3.13 x 10-7). Finally, based on the significance distribution of the associated microbiome QTLs in our study with respect to chromatin accessibility profiles, we identified tissues in which host genetic variation may be acting to influence bacterial abundance in the gut.
Conflict of interest statement
Figures
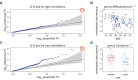

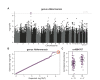
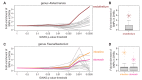
References
-
- Nadal I, Donat E, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. Journal of medical microbiology. 2007;56(Pt 12):1669–74. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

