Plants Release Precursors of Histone Deacetylase Inhibitors to Suppress Growth of Competitors
- PMID: 26530086
- PMCID: PMC4682303
- DOI: 10.1105/tpc.15.00585
Plants Release Precursors of Histone Deacetylase Inhibitors to Suppress Growth of Competitors
Abstract
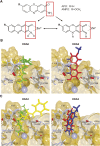
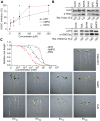
To secure their access to water, light, and nutrients, many plant species have developed allelopathic strategies to suppress competitors. To this end, they release into the rhizosphere phytotoxic substances that inhibit the germination and growth of neighbors. Despite the importance of allelopathy in shaping natural plant communities and for agricultural production, the underlying molecular mechanisms are largely unknown. Here, we report that allelochemicals derived from the common class of cyclic hydroxamic acid root exudates directly affect the chromatin-modifying machinery in Arabidopsis thaliana. These allelochemicals inhibit histone deacetylases both in vitro and in vivo and exert their activity through locus-specific alterations of histone acetylation and associated gene expression. Our multilevel analysis collectively shows how plant-plant interactions interfere with a fundamental cellular process, histone acetylation, by targeting an evolutionarily highly conserved class of enzymes.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures



References
-
- Anzai K., Isono K., Okuma K., Suzuki S. (1960). The new antibiotics, questiomycins A and B. J. Antibiot. 13: 125–132. - PubMed
-
- Baerson S.R., Sánchez-Moreiras A., Pedrol-Bonjoch N., Schulz M., Kagan I.A., Agarwal A.K., Reigosa M.J., Duke S.O. (2005). Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280: 21867–21881. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

