FILAMENTOUS FLOWER Is a Direct Target of JAZ3 and Modulates Responses to Jasmonate
- PMID: 26530088
- PMCID: PMC4682293
- DOI: 10.1105/tpc.15.00220
FILAMENTOUS FLOWER Is a Direct Target of JAZ3 and Modulates Responses to Jasmonate
Abstract
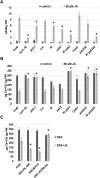
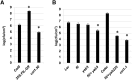
The plant hormone jasmonate (JA) plays an important role in regulating growth, development, and immunity. Activation of the JA-signaling pathway is based on the hormone-triggered ubiquitination and removal of transcriptional repressors (JASMONATE-ZIM DOMAIN [JAZ] proteins) by an SCF receptor complex (SCF(COI1)/JAZ). This removal allows the rapid activation of transcription factors (TFs) triggering a multitude of downstream responses. Identification of TFs bound by the JAZ proteins is essential to better understand how the JA-signaling pathway modulates and integrates different responses. In this study, we found that the JAZ3 repressor physically interacts with the YABBY (YAB) family transcription factor FILAMENTOUS FLOWER (FIL)/YAB1. In Arabidopsis thaliana, FIL regulates developmental processes such as axial patterning and growth of lateral organs, shoot apical meristem activity, and inflorescence phyllotaxy. Phenotypic analysis of JA-regulated responses in loss- and gain-of-function FIL lines suggested that YABs function as transcriptional activators of JA-triggered responses. Moreover, we show that MYB75, a component of the WD-repeat/bHLH/MYB complex regulating anthocyanin production, is a direct transcriptional target of FIL. We propose that JAZ3 interacts with YABs to attenuate their transcriptional function. Upon perception of JA signal, degradation of JAZ3 by the SCF(COI1) complex releases YABs to activate a subset of JA-regulated genes in leaves leading to anthocyanin accumulation, chlorophyll loss, and reduced bacterial defense.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures






References
-
- Barto E.K., Cipollini D. (2005). Testing the optimal defense theory and the growth-differentiation balance hypothesis in Arabidopsis thaliana. Oecologia 146: 169–178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

