tDRmapper: challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data
- PMID: 26530785
- PMCID: PMC4632369
- DOI: 10.1186/s12859-015-0800-0
tDRmapper: challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data
Abstract
Background: Small RNA-sequencing has revealed the diversity and high abundance of small RNAs derived from tRNAs, referred to as tRNA-derived RNAs. However, at present, there is no standardized nomenclature and there are no methods for accurate annotation and quantification of these small RNAs. tRNA-derived RNAs have unique features that limit the utility of conventional alignment tools and quantification methods.
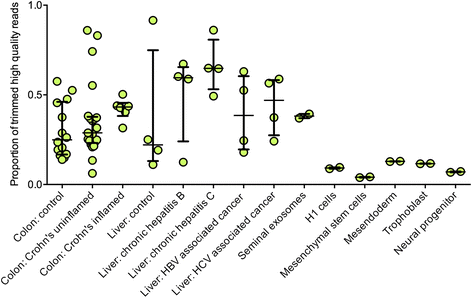
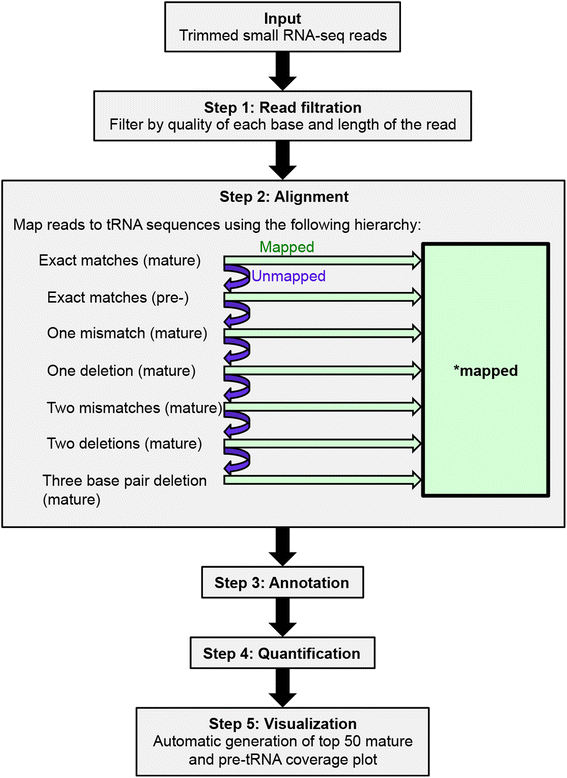
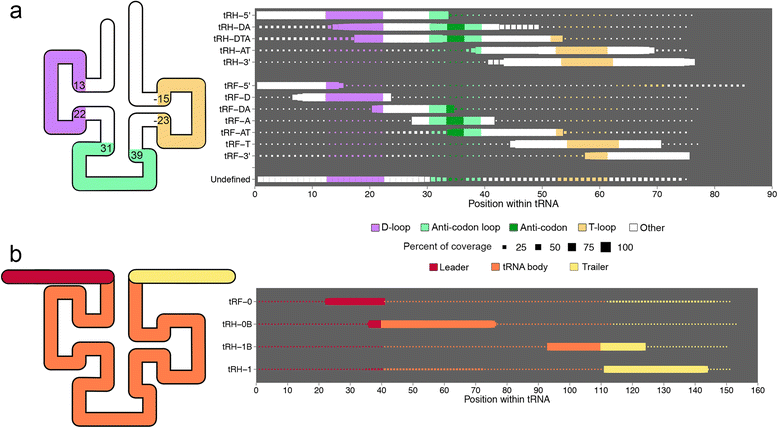
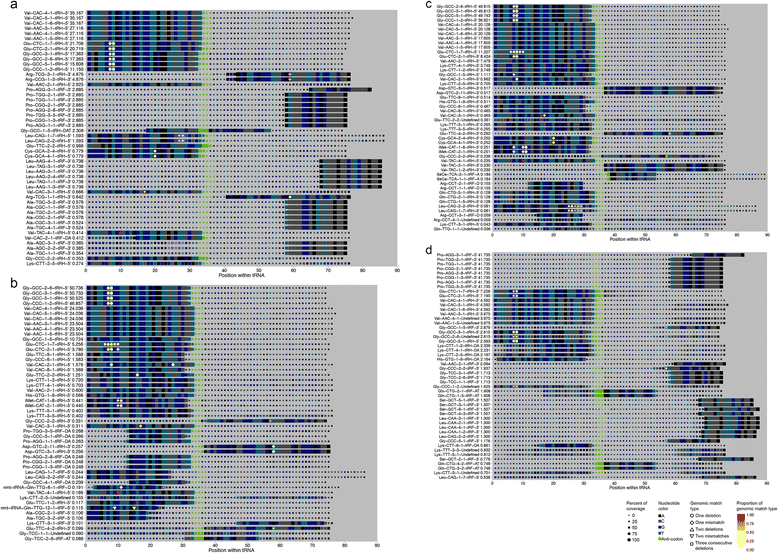
Results: We describe here the challenges of mapping, naming, and quantifying tRNA-derived RNAs and present a novel method that addresses them, called tDRmapper. We then use tDRmapper to perform a comparative analysis of tRNA-derived RNA profiles across different human cell types and diseases. We found that (1) tRNA-derived RNA profiles can differ dramatically across different cell types and disease states, (2) that positions and types of chemical modifications of tRNA-derived RNAs vary by cell type and disease, and (3) that entirely different tRNA-derived RNA species can be produced from the same parental tRNA depending on the cell type.
Conclusion: tDRmappernot only provides a standardized nomenclature and quantification scheme, but also includes graphical visualization that facilitates the discovery of novel tRNA and tRNA-derived RNA biology.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

