Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction
- PMID: 26531007
- PMCID: PMC4632034
- DOI: 10.1038/srep16076
Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction
Abstract
Achieving sufficient yields of proteins in their functional form represents the first bottleneck in contemporary bioscience and biotechnology. To accomplish successful overexpression of membrane proteins in a workhorse organism such as E. coli, defined and rational optimization strategies based on an understanding of the genetic background of the toxicity-escape mechanism are desirable. To this end, we sequenced the genomes of E. coli C41(DE3) and its derivative C43(DE3), which were developed for membrane protein production. Comparative analysis of their genomes with those of their ancestral strain E. coli BL21(DE3) revealed various genetic changes in both strains. A series of E. coli variants that are able to tolerate transformation with or overexpression of membrane proteins were generated by in vitro evolution. Targeted sequencing of the evolved strains revealed the mutational hotspots among the acquired genetic changes. By these combinatorial approaches, we found non-synonymous changes in the lac repressor gene of the lac operon as well as nucleotide substitutions in the lacUV5 promoter of the DE3 region, by which the toxic effect to the host caused by overexpression of membrane proteins could be relieved. A mutation in lacI was demonstrated to be crucial for conferring tolerance to membrane protein overexpression.
Figures

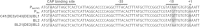



References
-
- Stevens T. J. & Arkin I. T. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins 39, 417–420 (2000). - PubMed
-
- White S. H. & Wimley W. C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1376, 339–352 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

