Whole exome sequencing (WES) on formalin-fixed, paraffin-embedded (FFPE) tumor tissue in gastrointestinal stromal tumors (GIST)
- PMID: 26531060
- PMCID: PMC4630927
- DOI: 10.1186/s12864-015-1982-6
Whole exome sequencing (WES) on formalin-fixed, paraffin-embedded (FFPE) tumor tissue in gastrointestinal stromal tumors (GIST)
Abstract
Background: Next generation sequencing (NGS) technology has been rapidly introduced into basic and translational research in oncology, but the reduced availability of fresh frozen (FF) tumor tissues and the poor quality of DNA extracted from formalin-fixed, paraffin-embedded (FFPE) has significantly impaired this process in the field of solid tumors. To evaluate if data generated from FFPE material can be reliably produced and potentially used in routine clinical settings, we performed whole exome sequencing (WES) from tumor samples of Gastrointestinal stromal tumors (GIST), either extracted FF or FFPE, and from matched normal DNA.
Methods: We performed whole exome enrichment and sequencing at 100bp in paired end on four GIST samples, either from FFPE or fresh-frozen tissue, and from matched normal DNA.
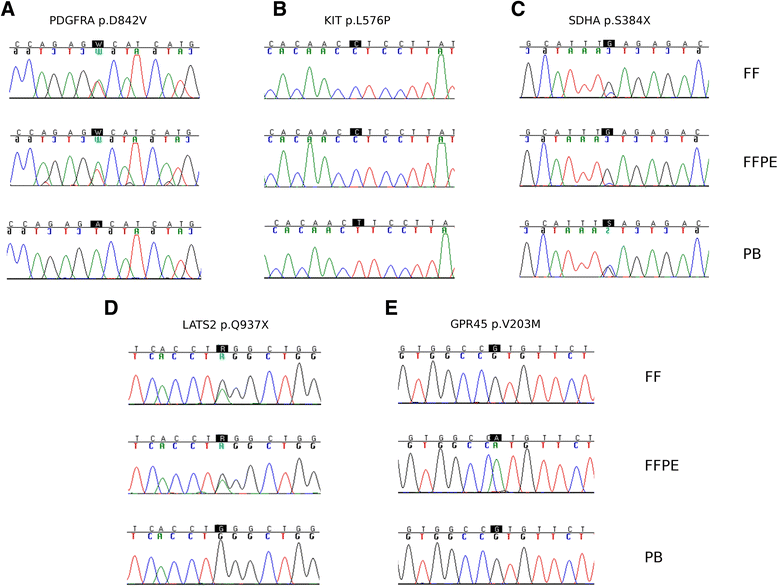
Results: The integrity of DNA extracted from FFPE was evaluated by a modified RAPD PCR method, thus identifying high quality (HQ) and low quality (LQ) FFPE. DNA library production and exome capture was feasible for both classes of FFPE, despite the smaller yield and insert size of LQ-FFPE. WES produced data of equal quality from FF and FFPE, while only HQ-FFPE yielded an amount of data comparable to FF samples. Bioinformatic analysis showed that the percentage of variants called both in FF and FFPE samples was very high in HQ-FFPE, reaching 94-96 % of the total number of called variants. Classification of somatic variants by nucleotide substitution type showed that HQ-FFPE and FF had similar mutational profiles, while LQ-FFPE samples carried a much higher number of mutations than the FF counterpart, with a significant enrichment of C > T/G > A substitutions. Focusing on potential disease-related variants allowed the discovery of additional somatic variants in GIST samples, apart from the known oncogenic driver mutation, both from sequencing of FF and FFPE material. False positive and false negative calls were present almost exclusively in the analysis of FFPE of low quality. On the whole this study showed that WES is feasible also on FFPE specimens and that it is possible to easily select FFPE samples of high quality that yield sequencing results comparable to the FF counterpart.
Conclusions: WES on FFPE material may represent an important and innovative source for GIST research and for other solid tumors, amenable of possible application in clinical practice.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

