Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica
- PMID: 26531864
- PMCID: PMC4632031
- DOI: 10.1038/srep16265
Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica
Abstract
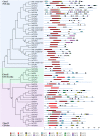
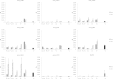
TCP proteins are plant-specific transcription factors involved in many different processes. Because of their involvement in a large number of developmental pathways, their roles have been investigated in various plant species. However, there are almost no studies of this transcription factor family in orchids. Based on the available transcriptome of the inflorescence of the orchid Orchis italica, in the present study we identified 12 transcripts encoding TCP proteins. The phylogenetic analysis showed that they belong to different TCP classes (I and II) and groups (PCF, CIN and CYC/TB1), and that they display a number of conserved motifs when compared with the TCPs of Arabidopsis and Oryza. The presence of a specific cleavage site for the microRNA miRNA319, an important post-transcriptional regulator of several TCP genes in other species, was demonstrated for one transcript of O. italica, and the analysis of the expression pattern of the TCP transcripts in different inflorescence organs and in leaf tissue suggests that some TCP transcripts of O. italica exert their role only in specific tissues, while others may play multiple roles in different tissues. In addition, the evolutionary analysis showed a general purifying selection acting on the coding region of these transcripts.
Figures



References
-
- Cubas P., Lauter N., Doebley J. & Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18, 215–222 (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

