Using Population Genetics to Interrogate the Monogenic Nephrotic Syndrome Diagnosis in a Case Cohort
- PMID: 26534921
- PMCID: PMC4926977
- DOI: 10.1681/ASN.2015050504
Using Population Genetics to Interrogate the Monogenic Nephrotic Syndrome Diagnosis in a Case Cohort
Abstract
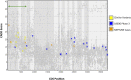
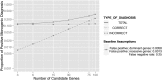
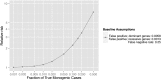
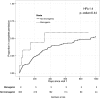
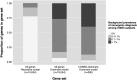
To maximize clinical benefits of genetic screening of patients with nephrotic syndrome (NS) to diagnose monogenic causes, reliably distinguishing NS-causing variants from the background of rare, noncausal variants prevalent in all genomes is vital. To determine the prevalence of monogenic NS in a North American case cohort while accounting for background prevalence of genetic variation, we sequenced 21 implicated monogenic NS genes in 312 participants from the Nephrotic Syndrome Study Network and 61 putative controls from the 1000 Genomes Project (1000G). These analyses were extended to available sequence data from approximately 2500 subjects from the 1000G. A typical pathogenicity filter identified causal variants for NS in 4.2% of patients and 5.8% of subjects from the 1000G. We devised a more stringent pathogenicity filtering strategy, reducing background prevalence of causal variants to 1.5%. When applying this stringent filter to patients, prevalence of monogenic NS was 2.9%; of these patients, 67% were pediatric, and 44% had FSGS on biopsy. The rate of complete remission did not associate with monogenic classification. Thus, we identified factors contributing to inaccurate monogenic classification of NS and developed a more accurate variant filtering strategy. The prevalence and clinical correlates of monogenic NS in this sporadically affected cohort differ substantially from those reported for patients referred for genetic analysis. Particularly in unselected, population-based cases, considering putative causal variants in known NS genes from a probabilistic rather than a deterministic perspective may be more precise. We also introduce GeneVetter, a web tool for monogenic assessment of rare disease.
Keywords: 1000 Genomes; expressivity; focal segmental glomerulosclerosis; genetic renal disease; penetrance; steroid resistant nephrotic syndrome.
Copyright © 2016 by the American Society of Nephrology.
Figures

References
-
- Saleem MA: New developments in steroid-resistant nephrotic syndrome. Pediatr Nephrol 28: 699–709, 2013 - PubMed
-
- Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Hildebrandt F SRNS Study Group : A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015 - PMC - PubMed
-
- McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA RADAR the UK SRNS Study Group : Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 8: 637–648, 2013 - PMC - PubMed
-
- Lipska BS, Iatropoulos P, Maranta R, Caridi G, Ozaltin F, Anarat A, Balat A, Gellermann J, Trautmann A, Erdogan O, Saeed B, Emre S, Bogdanovic R, Azocar M, Balasz-Chmielewska I, Benetti E, Caliskan S, Mir S, Melk A, Ertan P, Baskin E, Jardim H, Davitaia T, Wasilewska A, Drozdz D, Szczepanska M, Jankauskiene A, Higuita LM, Ardissino G, Ozkaya O, Kuzma-Mroczkowska E, Soylemezoglu O, Ranchin B, Medynska A, Tkaczyk M, Peco-Antic A, Akil I, Jarmolinski T, Firszt-Adamczyk A, Dusek J, Simonetti GD, Gok F, Gheissari A, Emma F, Krmar RT, Fischbach M, Printza N, Simkova E, Mele C, Ghiggeri GM, Schaefer F PodoNet Consortium : Genetic screening in adolescents with steroid-resistant nephrotic syndrome. Kidney Int 84: 206–213, 2013 - PubMed
-
- Giglio S, Provenzano A, Mazzinghi B, Becherucci F, Giunti L, Sansavini G, Ravaglia F, Roperto RM, Farsetti S, Benetti E, Rotondi M, Murer L, Lazzeri E, Lasagni L, Materassi M, Romagnani P: Heterogeneous genetic alterations in sporadic nephrotic syndrome associate with resistance to immunosuppression. J Am Soc Nephrol 26: 230–236, 2015 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

