Cleavage of Signal Regulatory Protein α (SIRPα) Enhances Inflammatory Signaling
- PMID: 26534964
- PMCID: PMC4692235
- DOI: 10.1074/jbc.M115.682914
Cleavage of Signal Regulatory Protein α (SIRPα) Enhances Inflammatory Signaling
Abstract
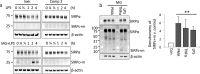
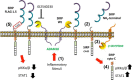
Signal regulatory protein α (SIRPα) is a membrane glycoprotein immunoreceptor abundant in cells of monocyte lineage. SIRPα ligation by a broadly expressed transmembrane protein, CD47, results in phosphorylation of the cytoplasmic immunoreceptor tyrosine-based inhibitory motifs, resulting in the inhibition of NF-κB signaling in macrophages. Here we observed that proteolysis of SIRPα during inflammation is regulated by a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10), resulting in the generation of a membrane-associated cleavage fragment in both THP-1 monocytes and human lung epithelia. We mapped a charge-dependent putative cleavage site near the membrane-proximal domain necessary for ADAM10-mediated cleavage. In addition, a secondary proteolytic cleavage within the membrane-associated SIRPα fragment by γ-secretase was identified. Ectopic expression of a SIRPα mutant plasmid encoding a proteolytically resistant form in HeLa cells inhibited activation of the NF-κB pathway and suppressed STAT1 phosphorylation in response to TNFα to a greater extent than expression of wild-type SIRPα. Conversely, overexpression of plasmids encoding the proteolytically cleaved SIRPα fragments in cells resulted in enhanced STAT-1 and NF-κB pathway activation. Thus, the data suggest that combinatorial actions of ADAM10 and γ-secretase on SIRPα cleavage promote inflammatory signaling.
Keywords: ADAM10; endotoxin; epithelium; inflammation; lung; protein degradation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Ravetch J. V., and Lanier L. L. (2000) Immune inhibitory receptors. Science 290, 84–89 - PubMed
-
- Steevels T. A., and Meyaard L. (2011) Immune inhibitory receptors: essential regulators of phagocyte function. Eur. J. Immunol. 41, 575–587 - PubMed
-
- Adams S., van der Laan L. J., Vernon-Wilson E., Renardel de Lavalette C., Döpp E. A., Dijkstra C. D., Simmons D. L., and van den Berg T. K. (1998) Signal-regulatory protein is selectively expressed by myeloid and neuronal cells. J. Immunol. 161, 1853–1859 - PubMed
-
- van Beek E. M., Cochrane F., Barclay A. N., and van den Berg T. K. (2005) Signal regulatory proteins in the immune system. J. Immunol. 175, 7781–7787 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- I01 BX002200/BX/BLRD VA/United States
- R01 HL097376/HL/NHLBI NIH HHS/United States
- R01 HL112914/HL/NHLBI NIH HHS/United States
- HL097376/HL/NHLBI NIH HHS/United States
- HL096376/HL/NHLBI NIH HHS/United States
- UH2 HL123502/HL/NHLBI NIH HHS/United States
- 1UH2HL123502/HL/NHLBI NIH HHS/United States
- 5T32HL007563-28/HL/NHLBI NIH HHS/United States
- P01 HL114453/HL/NHLBI NIH HHS/United States
- HL081784/HL/NHLBI NIH HHS/United States
- HL114453/HL/NHLBI NIH HHS/United States
- R01 HL096376/HL/NHLBI NIH HHS/United States
- HL098174/HL/NHLBI NIH HHS/United States
- R01 HL081784/HL/NHLBI NIH HHS/United States
- R01 HL098174/HL/NHLBI NIH HHS/United States
- T32 HL007563/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

