Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics
- PMID: 26535051
- PMCID: PMC4630929
- DOI: 10.1186/s13007-015-0093-3
Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics
Abstract
Background: Plant root systems are key drivers of plant function and yield. They are also under-explored targets to meet global food and energy demands. Many new technologies have been developed to characterize crop root system architecture (CRSA). These technologies have the potential to accelerate the progress in understanding the genetic control and environmental response of CRSA. Putting this potential into practice requires new methods and algorithms to analyze CRSA in digital images. Most prior approaches have solely focused on the estimation of root traits from images, yet no integrated platform exists that allows easy and intuitive access to trait extraction and analysis methods from images combined with storage solutions linked to metadata. Automated high-throughput phenotyping methods are increasingly used in laboratory-based efforts to link plant genotype with phenotype, whereas similar field-based studies remain predominantly manual low-throughput.
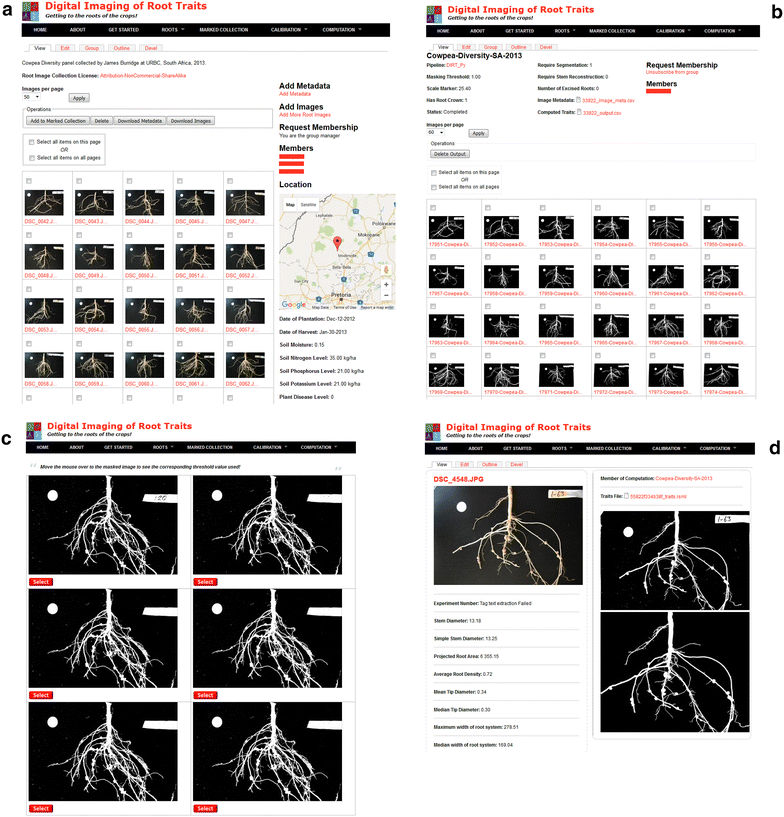
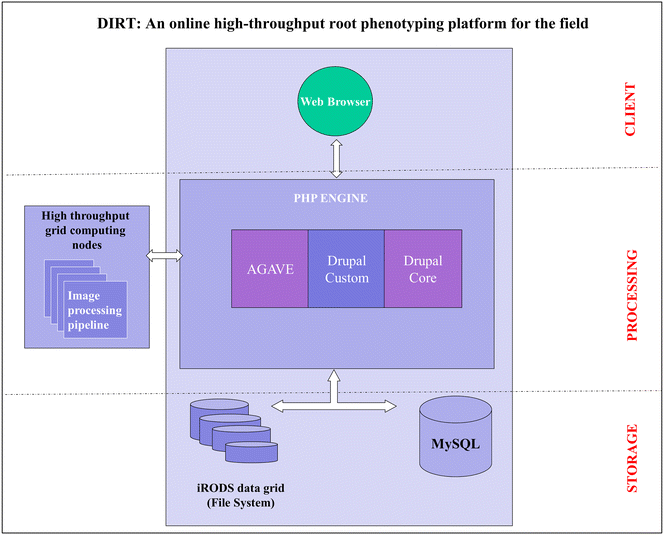
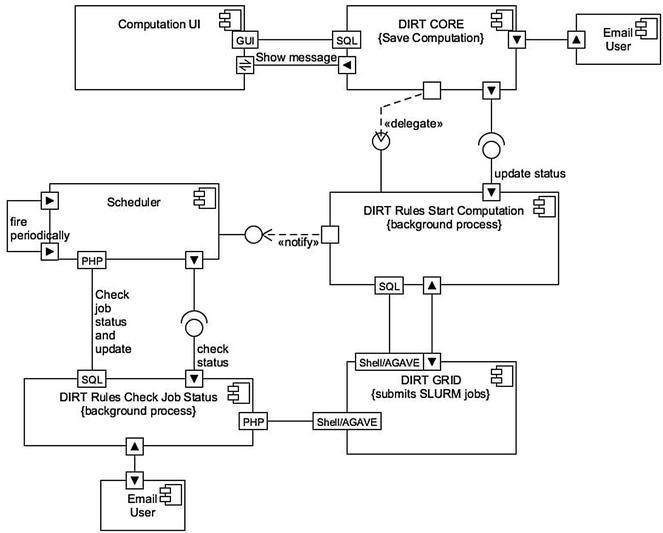
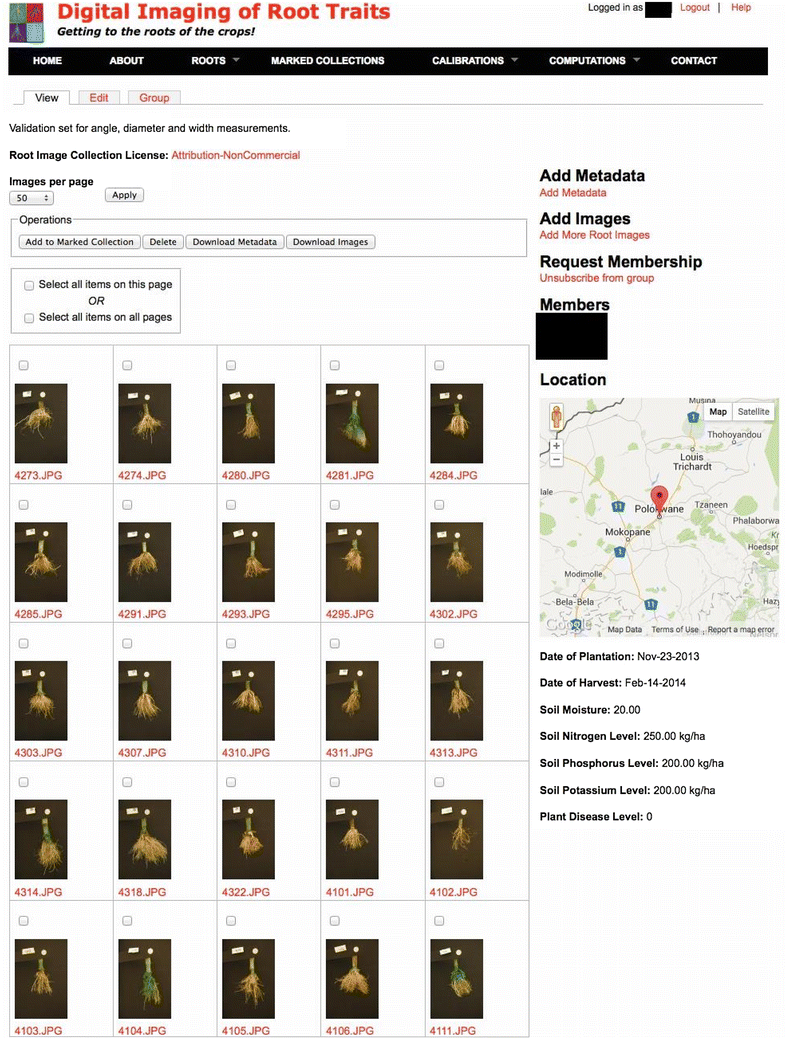
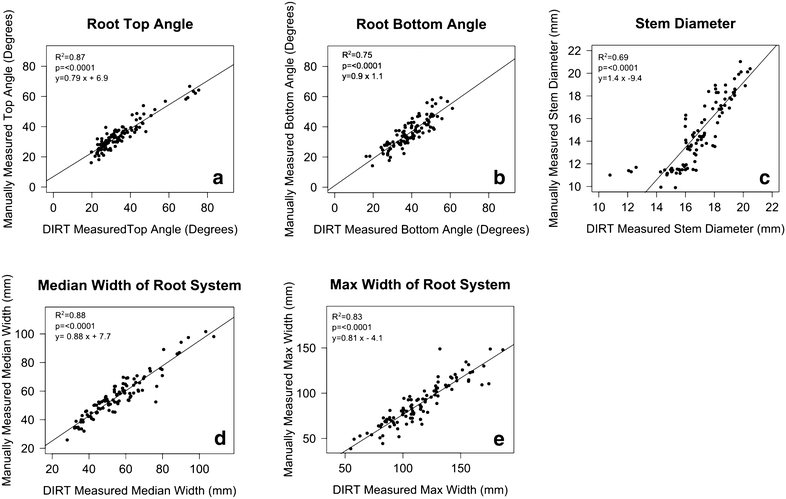
Description: Here, we present an open-source phenomics platform "DIRT", as a means to integrate scalable supercomputing architectures into field experiments and analysis pipelines. DIRT is an online platform that enables researchers to store images of plant roots, measure dicot and monocot root traits under field conditions, and share data and results within collaborative teams and the broader community. The DIRT platform seamlessly connects end-users with large-scale compute "commons" enabling the estimation and analysis of root phenotypes from field experiments of unprecedented size.
Conclusion: DIRT is an automated high-throughput computing and collaboration platform for field based crop root phenomics. The platform is accessible at http://www.dirt.iplantcollaborative.org/ and hosted on the iPlant cyber-infrastructure using high-throughput grid computing resources of the Texas Advanced Computing Center (TACC). DIRT is a high volume central depository and high-throughput RSA trait computation platform for plant scientists working on crop roots. It enables scientists to store, manage and share crop root images with metadata and compute RSA traits from thousands of images in parallel. It makes high-throughput RSA trait computation available to the community with just a few button clicks. As such it enables plant scientists to spend more time on science rather than on technology. All stored and computed data is easily accessible to the public and broader scientific community. We hope that easy data accessibility will attract new tool developers and spur creative data usage that may even be applied to other fields of science.
Figures

References
-
- OECD. OECD Environmental Outlook to 2030. OECD Publishing; 2008.
-
- Lynch JP. Roots of the second green revolution. Aust J Bot. 2007;55(5):493–512. doi: 10.1071/BT06118. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

