Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- PMID: 26536359
- PMCID: PMC4633059
- DOI: 10.1371/journal.pgen.1005641
Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
Abstract
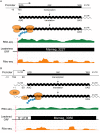
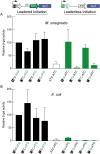
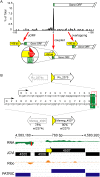
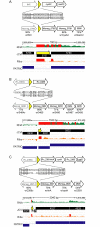
RNA-seq technologies have provided significant insight into the transcription networks of mycobacteria. However, such studies provide no definitive information on the translational landscape. Here, we use a combination of high-throughput transcriptome and proteome-profiling approaches to more rigorously understand protein expression in two mycobacterial species. RNA-seq and ribosome profiling in Mycobacterium smegmatis, and transcription start site (TSS) mapping and N-terminal peptide mass spectrometry in Mycobacterium tuberculosis, provide complementary, empirical datasets to examine the congruence of transcription and translation in the Mycobacterium genus. We find that nearly one-quarter of mycobacterial transcripts are leaderless, lacking a 5' untranslated region (UTR) and Shine-Dalgarno ribosome-binding site. Our data indicate that leaderless translation is a major feature of mycobacterial genomes and is comparably robust to leadered initiation. Using translational reporters to systematically probe the cis-sequence requirements of leaderless translation initiation in mycobacteria, we find that an ATG or GTG at the mRNA 5' end is both necessary and sufficient. This criterion, together with our ribosome occupancy data, suggests that mycobacteria encode hundreds of small, unannotated proteins at the 5' ends of transcripts. The conservation of small proteins in both mycobacterial species tested suggests that some play important roles in mycobacterial physiology. Our translational-reporter system further indicates that mycobacterial leadered translation initiation requires a Shine Dalgarno site in the 5' UTR and that ATG, GTG, TTG, and ATT codons can robustly initiate translation. Our combined approaches provide the first comprehensive view of mycobacterial gene structures and their non-canonical mechanisms of protein expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol. 1990;213(4):811–8. - PubMed
-
- Christie GE, Calendar R. Bacteriophage P2 late promoters. II. Comparison of the four late promoter sequences. Journal of molecular biology. 1985;181(3):373–82. Epub 1985/02/05. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

