HOXB9 induction of mesenchymal-to-epithelial transition in gastric carcinoma is negatively regulated by its hexapeptide motif
- PMID: 26536658
- PMCID: PMC4767475
- DOI: 10.18632/oncotarget.5814
HOXB9 induction of mesenchymal-to-epithelial transition in gastric carcinoma is negatively regulated by its hexapeptide motif
Abstract
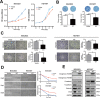
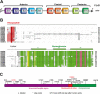
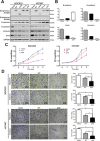
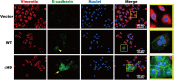
HOXB9, a transcription factor, plays an important role in development. While HOXB9 has been implicated in tumorigenesis and metastasis, its mechanisms are variable and its role in gastric carcinoma (GC) remains unclear. In the present study, we demonstrated that the expression of HOXB9 decreased in gastric carcinoma and was associated with malignancy and metastasis. Re-expression of HOXB9 in gastric cell lines resulted in the suppression of cell proliferation, migration, and invasion, which was accompanied by the induction of mesenchymal-to-epithelial transition (MET). Comparative sequence analysis and examination of a HOXB9 structural model indicated that three sites might possibly be involved in MET regulation. The in vitro study of HOXB9 mutants showed that these were unable to inhibit MET induction. However, when overexpressing a HOXB9 mutant lacking the hexapeptide motif, a more potent MET induction and tumor suppression was observed compared to that of the wild-type, indicating that the presence of the hexapeptide motif reduced HOXB9 MET induction and tumor suppression activity. Therefore, the results of the present study suggested that HOXB9 is a tumor suppressor in gastric carcinoma, and its activity was controlled by different regulatory mechanisms such as the hexapeptide motif as a "brake" in this case. The results of these regulatory effects could lead to either oncogenic or tumor suppressive roles of HOXB9, depending on the context of the particular type of cancer involved.
Keywords: HOXB9; gastric carcinoma; hexapeptide; mesenchymal-to-epithelial transition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annual review of biochemistry. 1994;63:487–526. - PubMed
-
- Apiou F, Flagiello D, Cillo C, Malfoy B, Poupon MF, Dutrillaux B. Fine mapping of human HOX gene clusters. Cytogenetics and cell genetics. 1996;73:114–115. - PubMed
-
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. - PubMed
-
- Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nature reviews Genetics. 2005;6:881–892. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

