Genetic interactions contribute less than additive effects to quantitative trait variation in yeast
- PMID: 26537231
- PMCID: PMC4635962
- DOI: 10.1038/ncomms9712
Genetic interactions contribute less than additive effects to quantitative trait variation in yeast
Abstract
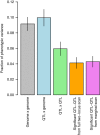
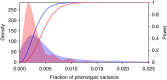
Genetic mapping studies of quantitative traits typically focus on detecting loci that contribute additively to trait variation. Genetic interactions are often proposed as a contributing factor to trait variation, but the relative contribution of interactions to trait variation is a subject of debate. Here we use a very large cross between two yeast strains to accurately estimate the fraction of phenotypic variance due to pairwise QTL-QTL interactions for 20 quantitative traits. We find that this fraction is 9% on average, substantially less than the contribution of additive QTL (43%). Statistically significant QTL-QTL pairs typically have small individual effect sizes, but collectively explain 40% of the pairwise interaction variance. We show that pairwise interaction variance is largely explained by pairs of loci at least one of which has a significant additive effect. These results refine our understanding of the genetic architecture of quantitative traits and help guide future mapping studies.
Figures


References
-
- Nelson R. M., Pettersson M. E. & Carlborg Ö. A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet. 29, 669–676 (2013). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

