Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles
- PMID: 26538482
- PMCID: PMC4633731
- DOI: 10.1038/srep16305
Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles
Abstract
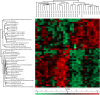
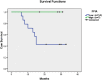
The development and progression of oral cavity squamous cell carcinoma (OSCC) involves complex cellular mechanisms that contribute to the low five-year survival rate of approximately 20% among diagnosed patients. However, the biological processes essential to tumor progression are not completely understood. Therefore, detecting alterations in the salivary proteome may assist in elucidating the cellular mechanisms modulated in OSCC and improve the clinical prognosis of the disease. The proteome of whole saliva and salivary extracellular vesicles (EVs) from patients with OSCC and healthy individuals were analyzed by LC-MS/MS and label-free protein quantification. Proteome data analysis was performed using statistical, machine learning and feature selection methods with additional functional annotation. Biological processes related to immune responses, peptidase inhibitor activity, iron coordination and protease binding were overrepresented in the group of differentially expressed proteins. Proteins related to the inflammatory system, transport of metals and cellular growth and proliferation were identified in the proteome of salivary EVs. The proteomics data were robust and could classify OSCC with 90% accuracy. The saliva proteome analysis revealed that immune processes are related to the presence of OSCC and indicate that proteomics data can contribute to determining OSCC prognosis.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

