Insights into the molecular mechanism of allostery in Hsp70s
- PMID: 26539440
- PMCID: PMC4611139
- DOI: 10.3389/fmolb.2015.00058
Insights into the molecular mechanism of allostery in Hsp70s
Abstract
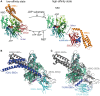
Hsp70s chaperone an amazing number and variety of cellular protein folding processes. Key to their versatility is the recognition of a short degenerate sequence motif, present in practically all polypeptides, and a bidirectional allosteric intramolecular regulation mechanism linking their N-terminal nucleotide binding domain (NBD) and their C-terminal polypeptide substrate binding domain (SBD). Through this interdomain communication ATP binding to the NBD and ATP hydrolysis control the affinity of the SBD for polypeptide substrates and substrate binding to the SBD triggers ATP hydrolysis. Genetic screens for defective variants of Hsp70s and systematic analysis of available structures of the isolated domains revealed some residues involved in allosteric control. Recent elucidation of the crystal structure of the Hsp70 homolog DnaK in the ATP bound open conformation as well as numerous NMR and mutagenesis studies bring us closer to an understanding of the communication between NBD and SBD. In this review we will discuss our current view of the allosteric control mechanism of Hsp70 chaperones.
Keywords: Hsp70 heat-shock proteins; allostery; conformational dynamics; interdomain communication; structure-function relationships.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

