Construction of the High-Density Genetic Linkage Map and Chromosome Map of Large Yellow Croaker (Larimichthys crocea)
- PMID: 26540048
- PMCID: PMC4661810
- DOI: 10.3390/ijms161125951
Construction of the High-Density Genetic Linkage Map and Chromosome Map of Large Yellow Croaker (Larimichthys crocea)
Abstract
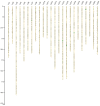
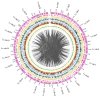
High-density genetic maps are essential for genome assembly, comparative genomic analysis and fine mapping of complex traits. In this study, 31,191 single nucleotide polymorphisms (SNPs) evenly distributed across the large yellow croaker (Larimichthys crocea) genome were identified using restriction-site associated DNA sequencing (RAD-seq). Among them, 10,150 high-confidence SNPs were assigned to 24 consensus linkage groups (LGs). The total length of the genetic linkage map was 5451.3 cM with an average distance of 0.54 cM between loci. This represents the densest genetic map currently reported for large yellow croaker. Using 2889 SNPs to target specific scaffolds, we assigned 533 scaffolds, comprising 421.44 Mb (62.04%) of the large yellow croaker assembled sequence, to the 24 linkage groups. The mapped assembly scaffolds in large yellow croaker were used for genome synteny analyses against the stickleback (Gasterosteus aculeatus) and medaka (Oryzias latipes). Greater synteny was observed between large yellow croaker and stickleback. This supports the hypothesis that large yellow croaker is more closely related to stickleback than to medaka. Moreover, 1274 immunity-related genes and 195 hypoxia-related genes were mapped to the 24 chromosomes of large yellow croaker. The integration of the high-resolution genetic map and the assembled sequence provides a valuable resource for fine mapping and positional cloning of quantitative trait loci associated with economically important traits in large yellow croaker.
Keywords: SNP; chromosome map; genetic linkage map; hypoxia-related genes; immunity-related genes; large yellow croaker (Larimichthys crocea).
Figures

References
-
- Feng Z., Cao Q. Ichthyology. 1st ed. Agricultural Press House; Beijing, China: 1979. pp. 154–155.
-
- Ning Y., Liu X., Wang Z.Y., Guo W., Li Y., Xie F. A genetic map of large yellow croaker pseudosciaena crocea. Aquaculture. 2007;264:16–26. doi: 10.1016/j.aquaculture.2006.12.042. - DOI
-
- Su Y. Breeding and Farming of Pseudosciaena Crocea. 1st ed. China Ocean Press; Beijing, China: 2004. pp. 1–68.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

