Redefining the BH3 Death Domain as a 'Short Linear Motif'
- PMID: 26541461
- PMCID: PMC5056427
- DOI: 10.1016/j.tibs.2015.09.007
Redefining the BH3 Death Domain as a 'Short Linear Motif'
Abstract
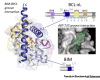
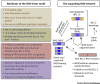
B cell lymphoma-2 (BCL-2)-related proteins control programmed cell death through a complex network of protein-protein interactions mediated by BCL-2 homology 3 (BH3) domains. Given their roles as dynamic linchpins, the discovery of novel BH3-containing proteins has attracted considerable attention. However, without a clearly defined BH3 signature sequence the BCL-2 family has expanded to include a nebulous group of nonhomologous BH3-only proteins, now justified by an intriguing twist. We present evidence that BH3s from both ordered and disordered proteins represent a new class of short linear motifs (SLiMs) or molecular recognition features (MoRFs) and are diverse in their evolutionary histories. The implied corollaries are that BH3s have a broad phylogenetic distribution and could potentially bind to non-BCL-2-like structural domains with distinct functions.
Keywords: BCL-2 family; BH3; MoRF/MoRE; SLiM; globular domains; intrinsically disordered proteins.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

References
-
- Czabotar P.E. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. - PubMed
-
- Aouacheria A. Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-only, and BNip families of apoptotic regulators. Mol. Biol. Evol. 2005;22:2395–2416. - PubMed
-
- Kvansakul M., Hinds M.G. The structural biology of BH3-only proteins. Methods Enzymol. 2014;544:49–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

