Distinct routes of lineage development reshape the human blood hierarchy across ontogeny
- PMID: 26541609
- PMCID: PMC4816201
- DOI: 10.1126/science.aab2116
Distinct routes of lineage development reshape the human blood hierarchy across ontogeny
Abstract
In a classical view of hematopoiesis, the various blood cell lineages arise via a hierarchical scheme starting with multipotent stem cells that become increasingly restricted in their differentiation potential through oligopotent and then unipotent progenitors. We developed a cell-sorting scheme to resolve myeloid (My), erythroid (Er), and megakaryocytic (Mk) fates from single CD34(+) cells and then mapped the progenitor hierarchy across human development. Fetal liver contained large numbers of distinct oligopotent progenitors with intermingled My, Er, and Mk fates. However, few oligopotent progenitor intermediates were present in the adult bone marrow. Instead, only two progenitor classes predominate, multipotent and unipotent, with Er-Mk lineages emerging from multipotent cells. The developmental shift to an adult "two-tier" hierarchy challenges current dogma and provides a revised framework to understand normal and disease states of human hematopoiesis.
Copyright © 2016, American Association for the Advancement of Science.
Figures
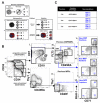


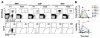



Comment in
-
STEM CELLS. Potency finds its niches.Science. 2016 Jan 8;351(6269):126-7. doi: 10.1126/science.aae0325. Science. 2016. PMID: 26744396 No abstract available.
-
On the origin of blood cells--hematopoiesis revisited.Oral Dis. 2016 May;22(4):247-8. doi: 10.1111/odi.12445. Epub 2016 Feb 15. Oral Dis. 2016. PMID: 26802784 Free PMC article. No abstract available.
References
-
- TILL JE, McCULLOCH EA. Hemopoietic stem cell differentiation. Biochim. Biophys. Acta. 1980;605:431–459. - PubMed
-
- Spangrude GJ, et al. Mouse hematopoietic stem cells. Blood. 1991;78:1395–1402. - PubMed
-
- Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. 2005;121:295–306. - PubMed
-
- Månsson R, et al. Molecular Evidence for Hierarchical Transcriptional Lineage Priming in Fetal and Adult Stem Cells and Multipotent Progenitors. Immunity. 2007;26:407–419. - PubMed
-
- Yamamoto R, et al. Clonal Analysis Unveils Self-Renewing Lineage-Restricted Progenitors Generated Directly from Hematopoietic Stem Cells. 2013;154:1112–1126. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

