Giant peroxisomes in a moss (Physcomitrella patens) peroxisomal biogenesis factor 11 mutant
- PMID: 26542980
- PMCID: PMC4738463
- DOI: 10.1111/nph.13739
Giant peroxisomes in a moss (Physcomitrella patens) peroxisomal biogenesis factor 11 mutant
Abstract
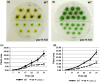
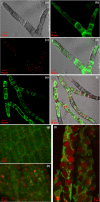
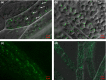
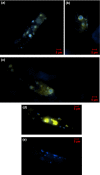
Peroxisomal biogenesis factor 11 (PEX11) proteins are found in yeasts, mammals and plants, and play a role in peroxisome morphology and regulation of peroxisome division. The moss Physcomitrella patens has six PEX11 isoforms which fall into two subfamilies, similar to those found in monocots and dicots. We carried out targeted gene disruption of the Phypa_PEX11-1 gene and compared the morphological and cellular phenotypes of the wild-type and mutant strains. The mutant grew more slowly and the development of gametophores was retarded. Mutant chloronemal filaments contained large cellular structures which excluded all other cellular organelles. Expression of fluorescent reporter proteins revealed that the mutant strain had greatly enlarged peroxisomes up to 10 μm in diameter. Expression of a vacuolar membrane marker confirmed that the enlarged structures were not vacuoles, or peroxisomes sequestered within vacuoles as a result of pexophagy. Phypa_PEX11 targeted to peroxisome membranes could rescue the knock out phenotype and interacted with Fission1 on the peroxisome membrane. Moss PEX11 functions in peroxisome division similar to PEX11 in other organisms but the mutant phenotype is more extreme and environmentally determined, making P. patens a powerful system in which to address mechanisms of peroxisome proliferation and division.
Keywords: Physcomitrella patens; gene disruption; organelle size; peroxisomal biogenesis factor 11 (PEX11); peroxisome.
© 2015 The Authors. New Phytologist © 2015 New Phytologist Trust.
Figures




Similar articles
-
The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis.Plant Cell. 2007 Jan;19(1):333-50. doi: 10.1105/tpc.106.045831. Epub 2007 Jan 12. Plant Cell. 2007. PMID: 17220199 Free PMC article.
-
Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae.Mol Biol Cell. 2003 Oct;14(10):4089-102. doi: 10.1091/mbc.e03-03-0150. Epub 2003 May 18. Mol Biol Cell. 2003. PMID: 14517321 Free PMC article.
-
A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes.Traffic. 2012 Jan;13(1):157-67. doi: 10.1111/j.1600-0854.2011.01290.x. Epub 2011 Oct 20. Traffic. 2012. PMID: 21951626 Free PMC article.
-
Peroxisome biogenesis and inter-organelle communication: an indispensable role for Pex11 and Pex30 family proteins in yeast.Curr Genet. 2022 Dec;68(5-6):537-550. doi: 10.1007/s00294-022-01254-y. Epub 2022 Oct 15. Curr Genet. 2022. PMID: 36242632 Review.
-
Yeast peroxisomes: How are they formed and how do they grow?Int J Biochem Cell Biol. 2018 Dec;105:24-34. doi: 10.1016/j.biocel.2018.09.019. Epub 2018 Sep 27. Int J Biochem Cell Biol. 2018. PMID: 30268746 Review.
Cited by
-
Towards designer organelles by subverting the peroxisomal import pathway.Nat Commun. 2017 Sep 6;8(1):454. doi: 10.1038/s41467-017-00487-7. Nat Commun. 2017. PMID: 28878206 Free PMC article.
-
Conserved and differential transcriptional responses of peroxisome associated pathways to drought, dehydration and ABA.J Exp Bot. 2018 Sep 14;69(20):4971-4985. doi: 10.1093/jxb/ery266. J Exp Bot. 2018. PMID: 30032264 Free PMC article.
-
Peroxisome Function, Biogenesis, and Dynamics in Plants.Plant Physiol. 2018 Jan;176(1):162-177. doi: 10.1104/pp.17.01050. Epub 2017 Oct 11. Plant Physiol. 2018. PMID: 29021223 Free PMC article. Review.
-
Transfection of Arctic Bryum sp. KMR5045 as a Model for Genetic Engineering of Cold-Tolerant Mosses.Front Plant Sci. 2021 Jan 8;11:609847. doi: 10.3389/fpls.2020.609847. eCollection 2020. Front Plant Sci. 2021. PMID: 33584753 Free PMC article.
References
-
- Abramoff M, Magalhaes P, Ram S. 2004. Processing with ImageJ. Biophotonics International 11: 36–42.
-
- Chang J, Klute MJ, Tower RJ, Mast FD, Dacks JB, Rachubinski RA. 2015. An ancestral role in peroxisome assembly is retained by the divisional peroxin Pex11 in the yeast Yarrowia lipolytica . Journal of Cell Science 128: 1327–1340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

