A Tandem Oligonucleotide Approach for SNP-Selective RNA Degradation Using Modified Antisense Oligonucleotides
- PMID: 26544037
- PMCID: PMC4704561
- DOI: 10.1371/journal.pone.0142139
A Tandem Oligonucleotide Approach for SNP-Selective RNA Degradation Using Modified Antisense Oligonucleotides
Erratum in
-
Correction: A Tandem Oligonucleotide Approach for SNP-Selective RNA Degradation Using Modified Antisense Oligonucleotides.PLoS One. 2016 Sep 19;11(9):e0163575. doi: 10.1371/journal.pone.0163575. eCollection 2016. PLoS One. 2016. PMID: 27643993 Free PMC article.
Abstract
Antisense oligonucleotides have been studied for many years as a tool for gene silencing. One of the most difficult cases of selective RNA silencing involves the alleles of single nucleotide polymorphisms, in which the allele sequence is differentiated by a single nucleotide. A new approach to improve the performance of allele selectivity for antisense oligonucleotides is proposed. It is based on the simultaneous application of two oligonucleotides. One is complementary to the mutated form of the targeted RNA and is able to activate RNase H to cleave the RNA. The other oligonucleotide, which is complementary to the wild type allele of the targeted RNA, is able to inhibit RNase H cleavage. Five types of SNPs, C/G, G/C, G/A, A/G, and C/U, were analyzed within the sequence context of genes associated with neurodegenerative disorders such as Alzheimer's disease, Parkinson's disease, ALS (Amyotrophic Lateral Sclerosis), and Machado-Joseph disease. For most analyzed cases, the application of the tandem approach increased allele-selective RNA degradation 1.5-15 fold relative to the use of a single antisense oligonucleotide. The presented study proves that differentiation between single substitution is highly dependent on the nature of the SNP and surrounding nucleotides. These variables are crucial for determining the proper length of the inhibitor antisense oligonucleotide. In the tandem approach, the comparison of thermodynamic stability of the favorable duplexes WT RNA-inhibitor and Mut RNA-gapmer with the other possible duplexes allows for the evaluation of chances for the allele-selective degradation of RNA. A larger difference in thermodynamic stability between favorable duplexes and those that could possibly form, usually results in the better allele selectivity of RNA degradation.
Conflict of interest statement
Figures
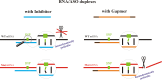
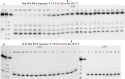


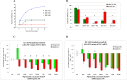

Similar articles
-
How RNase HI (Escherichia coli) promoted site-selective hydrolysis works on RNA in duplex with carba-LNA and LNA substituted antisense strands in an antisense strategy context?Mol Biosyst. 2017 May 2;13(5):921-938. doi: 10.1039/c6mb00762g. Mol Biosyst. 2017. PMID: 28352859
-
Differential effects on allele selective silencing of mutant huntingtin by two stereoisomers of α,β-constrained nucleic acid.ACS Chem Biol. 2014 Sep 19;9(9):1975-9. doi: 10.1021/cb5003027. Epub 2014 Jul 25. ACS Chem Biol. 2014. PMID: 25050989
-
Binding affinity and specificity of Escherichia coli RNase H1: impact on the kinetics of catalysis of antisense oligonucleotide-RNA hybrids.Biochemistry. 1997 Jan 14;36(2):390-8. doi: 10.1021/bi962230p. Biochemistry. 1997. PMID: 9003192
-
Is irrelevant cleavage the price of antisense efficacy?Pharmacol Ther. 2000 Mar;85(3):231-6. doi: 10.1016/s0163-7258(99)00053-4. Pharmacol Ther. 2000. PMID: 10739877 Review.
-
2'-Modified oligonucleotides for antisense therapeutics.Curr Top Med Chem. 2007;7(7):641-9. doi: 10.2174/156802607780487713. Curr Top Med Chem. 2007. PMID: 17430205 Review.
Cited by
-
Correction: A Tandem Oligonucleotide Approach for SNP-Selective RNA Degradation Using Modified Antisense Oligonucleotides.PLoS One. 2016 Sep 19;11(9):e0163575. doi: 10.1371/journal.pone.0163575. eCollection 2016. PLoS One. 2016. PMID: 27643993 Free PMC article.
-
A systematic study on the influence of thermodynamic asymmetry of 5'-ends of siRNA duplexes in relation to their silencing potency.Sci Rep. 2019 Feb 21;9(1):2477. doi: 10.1038/s41598-018-36620-9. Sci Rep. 2019. PMID: 30792489 Free PMC article.
-
Modified RNA triplexes: Thermodynamics, structure and biological potential.Sci Rep. 2018 Aug 29;8(1):13023. doi: 10.1038/s41598-018-31387-5. Sci Rep. 2018. PMID: 30158667 Free PMC article.
-
The oligonucleotides containing N7-regioisomer of guanosine: influence on thermodynamic properties and structure of RNA duplexes.RNA. 2024 Dec 16;31(1):86-99. doi: 10.1261/rna.080106.124. RNA. 2024. PMID: 39467648 Free PMC article.
-
Allele-Selective Thiomorpholino Antisense Oligonucleotides as a Therapeutic Approach for Fused-in-Sarcoma Amyotrophic Lateral Sclerosis.Int J Mol Sci. 2024 Aug 3;25(15):8495. doi: 10.3390/ijms25158495. Int J Mol Sci. 2024. PMID: 39126066 Free PMC article.
References
-
- Stull RA, Szoka FC Jr. Antigene, ribozyme and aptamer nucleic acid drugs: progress and prospects. Pharm Res. 1995;12(4):465–83. Epub 1995/04/01. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

