Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers
- PMID: 26546618
- PMCID: PMC4794379
- DOI: 10.1158/1078-0432.CCR-15-1259
Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers
Abstract
Purpose: Morphologically heterogeneous prostate cancers that behave clinically like small-cell prostate cancers (SCPC) share their chemotherapy responsiveness. We asked whether these clinically defined, morphologically diverse, "aggressive variant prostate cancer (AVPC)" also share molecular features with SCPC.
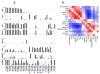
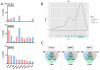
Experimental design: Fifty-nine prostate cancer samples from 40 clinical trial participants meeting AVPC criteria, and 8 patient-tumor derived xenografts (PDX) from 6 of them, were stained for markers aberrantly expressed in SCPC. DNA from 36 and 8 PDX was analyzed by Oncoscan for copy number gains (CNG) and losses (CNL). We used the AVPC PDX to expand observations and referenced publicly available datasets to arrive at a candidate molecular signature for the AVPC.
Results: Irrespective of morphology, Ki67 and Tp53 stained ≥10% cells in 80% and 41% of samples, respectively. RB1 stained <10% cells in 61% of samples and AR in 36%. MYC (surrogate for 8q) CNG and RB1 CNL showed in 54% of 44 samples each and PTEN CNL in 48%. All but 1 of 8 PDX bore Tp53 missense mutations. RB1 CNL was the strongest discriminator between unselected castration-resistant prostate cancer (CRPC) and the AVPC. Combined alterations in RB1, Tp53, and/or PTEN were more frequent in the AVPC than in unselected CRPC and in The Cancer Genome Atlas samples.
Conclusions: Clinically defined AVPC share molecular features with SCPC and are characterized by combined alterations in RB1, Tp53, and/or PTEN.
©2015 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest: None
Figures


References
-
- Jensen EV, Block GE, Smith S, Kyser K, DeSombre ER. Estrogen receptors and breast cancer response to adrenalectomy. National Cancer Institute monograph. 1971;34:55–70. - PubMed
-
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350(21):2129–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

