Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster
- PMID: 26547394
- PMCID: PMC5089930
- DOI: 10.1111/mec.13455
Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster
Abstract
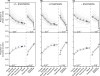
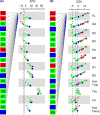
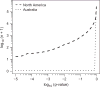
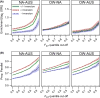
Populations arrayed along broad latitudinal gradients often show patterns of clinal variation in phenotype and genotype. Such population differentiation can be generated and maintained by both historical demographic events and local adaptation. These evolutionary forces are not mutually exclusive and can in some cases produce nearly identical patterns of genetic differentiation among populations. Here, we investigate the evolutionary forces that generated and maintain clinal variation genome-wide among populations of Drosophila melanogaster sampled in North America and Australia. We contrast patterns of clinal variation in these continents with patterns of differentiation among ancestral European and African populations. Using established and novel methods we derive here, we show that recently derived North America and Australia populations were likely founded by both European and African lineages and that this hybridization event likely contributed to genome-wide patterns of parallel clinal variation between continents. The pervasive effects of admixture mean that differentiation at only several hundred loci can be attributed to the operation of spatially varying selection using an FST outlier approach. Our results provide novel insight into the well-studied system of clinal differentiation in D. melanogaster and provide a context for future studies seeking to identify loci contributing to local adaptation in a wide variety of organisms, including other invasive species as well as temperate endemics.
Keywords: Drosophila melanogaster; adaptation; latitudinal clines; parallelism; secondary contact.
© 2015 John Wiley & Sons Ltd.
Figures
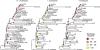
Comment in
-
Genomics of clinal variation in Drosophila: disentangling the interactions of selection and demography.Mol Ecol. 2016 Mar;25(5):1023-6. doi: 10.1111/mec.13534. Mol Ecol. 2016. PMID: 26919307
References
-
- Adams SM, Lindmeier JB, Duvernell DD. Microsatellite analysis of the phylogeography, Pleistocene history and secondary contact hypotheses for the killifish, Fundulus heteroclitus. Molecular Ecology. 2006;15:1109–1123. - PubMed
-
- Agosta SJ, Klemens JA. Ecological fitting by phenotypically flexible genotypes: implications for species associations, community assembly and evolution. Ecology Letters. 2008;11:1123–1134. - PubMed
-
- Andolfatto P. Contrasting Patterns of X-Linked and Autosomal Nucleotide Variation in Drosophila melanogaster and Drosophila simulans. Molecular Biology and Evolution. 2001;18:279–290. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

