The scaffold protein KSR1, a novel therapeutic target for the treatment of Merlin-deficient tumors
- PMID: 26549023
- PMCID: PMC4861249
- DOI: 10.1038/onc.2015.404
The scaffold protein KSR1, a novel therapeutic target for the treatment of Merlin-deficient tumors
Abstract
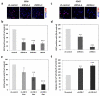
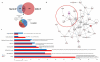
Merlin has broad tumor-suppressor functions as its mutations have been identified in multiple benign tumors and malignant cancers. In all schwannomas, the majority of meningiomas and 1/3 of ependymomas Merlin loss is causative. In neurofibromatosis type 2, a dominantly inherited tumor disease because of the loss of Merlin, patients suffer from multiple nervous system tumors and die on average around age 40. Chemotherapy is not effective and tumor localization and multiplicity make surgery and radiosurgery challenging and morbidity is often considerable. Thus, a new therapeutic approach is needed for these tumors. Using a primary human in vitro model for Merlin-deficient tumors, we report that the Ras/Raf/mitogen-activated protein, extracellular signal-regulated kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) scaffold, kinase suppressor of Ras 1 (KSR1), has a vital role in promoting schwannomas development. We show that KSR1 overexpression is involved in many pathological phenotypes caused by Merlin loss, namely multipolar morphology, enhanced cell-matrix adhesion, focal adhesion and, most importantly, increased proliferation and survival. Our data demonstrate that KSR1 has a wider role than MEK1/2 in the development of schwannomas because adhesion is more dependent on KSR1 than MEK1/2. Immunoprecipitation analysis reveals that KSR1 is a novel binding partner of Merlin, which suppresses KSR1's function by inhibiting the binding between KSR1 and c-Raf. Our proteomic analysis also demonstrates that KSR1 interacts with several Merlin downstream effectors, including E3 ubiquitin ligase CRL4(DCAF1). Further functional studies suggests that KSR1 and DCAF1 may co-operate to regulate schwannomas formation. Taken together, these findings suggest that KSR1 serves as a potential therapeutic target for Merlin-deficient tumors.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

