Rational design of a monomeric and photostable far-red fluorescent protein for fluorescence imaging in vivo
- PMID: 26549191
- PMCID: PMC4815332
- DOI: 10.1002/pro.2843
Rational design of a monomeric and photostable far-red fluorescent protein for fluorescence imaging in vivo
Abstract
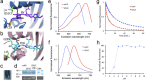
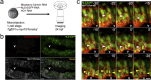
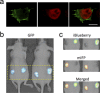
Fluorescent proteins (FPs) are powerful tools for cell and molecular biology. Here based on structural analysis, a blue-shifted mutant of a recently engineered monomeric infrared fluorescent protein (mIFP) has been rationally designed. This variant, named iBlueberry, bears a single mutation that shifts both excitation and emission spectra by approximately 40 nm. Furthermore, iBlueberry is four times more photostable than mIFP, rendering it more advantageous for imaging protein dynamics. By tagging iBlueberry to centrin, it has been demonstrated that the fusion protein labels the centrosome in the developing zebrafish embryo. Together with GFP-labeled nucleus and tdTomato-labeled plasma membrane, time-lapse imaging to visualize the dynamics of centrosomes in radial glia neural progenitors in the intact zebrafish brain has been demonstrated. It is further shown that iBlueberry can be used together with mIFP in two-color protein labeling in living cells and in two-color tumor labeling in mice.
Keywords: bacterial phytochrome; fluorescence imaging; fluorescent proteins; rational design.
© 2015 The Protein Society.
Figures
References
-
- Tsien RY (2009) Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture). Angew Chem Int Ed Engl 48:5612–5626. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

