Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2
- PMID: 26549758
- PMCID: PMC4667618
- DOI: 10.1038/ncomms9823
Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2
Abstract
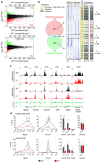
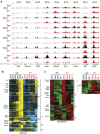
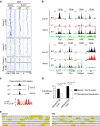
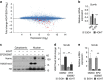
T-cell development is accompanied by epigenetic changes that ensure the silencing of stem cell-related genes and the activation of lymphocyte-specific programmes. How transcription factors influence these changes remains unclear. We show that the Ikaros transcription factor forms a complex with Polycomb repressive complex 2 (PRC2) in CD4(-)CD8(-) thymocytes and allows its binding to more than 500 developmentally regulated loci, including those normally activated in haematopoietic stem cells and others induced by the Notch pathway. Loss of Ikaros in CD4(-)CD8(-) cells leads to reduced histone H3 lysine 27 trimethylation and ectopic gene expression. Furthermore, Ikaros binding triggers PRC2 recruitment and Ikaros interacts with PRC2 independently of the nucleosome remodelling and deacetylation complex. Our results identify Ikaros as a fundamental regulator of PRC2 function in developing T cells.
Figures



References
-
- Vigano M. A. et al.. An epigenetic profile of early T-cell development from multipotent progenitors to committed T-cell descendants. Eur. J. Immunol. 44, 1181–1193 (2014). - PubMed
-
- Geimer Le Lay A. S. et al.. The tumor suppressor Ikaros shapes the repertoire of Notch target genes in T cells. Sci. Signal. 7, ra28 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

