Revisiting the reference genomes of human pathogenic Cryptosporidium species: reannotation of C. parvum Iowa and a new C. hominis reference
- PMID: 26549794
- PMCID: PMC4637869
- DOI: 10.1038/srep16324
Revisiting the reference genomes of human pathogenic Cryptosporidium species: reannotation of C. parvum Iowa and a new C. hominis reference
Abstract
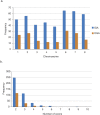
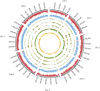
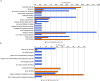
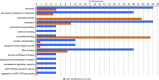
Cryptosporidium parvum and C. hominis are the most relevant species of this genus for human health. Both cause a self-limiting diarrhea in immunocompetent individuals, but cause potentially life-threatening disease in the immunocompromised. Despite the importance of these pathogens, only one reference genome of each has been analyzed and published. These two reference genomes were sequenced using automated capillary sequencing; as of yet, no next generation sequencing technology has been applied to improve their assemblies and annotations. For C. hominis, the main challenge that prevents a larger number of genomes to be sequenced is its resistance to axenic culture. In the present study, we employed next generation technology to analyse the genomic DNA and RNA to generate a new reference genome sequence of a C. hominis strain isolated directly from human stool and a new genome annotation of the C. parvum Iowa reference genome.
Figures

References
-
- Ryan U., Fayer R. & Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141, 1667–85 (2014). - PubMed
-
- Morgan U. M. et al. Cryptosporidium hominis n. sp.(Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49, 433–440 (2002). - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

