Full-length single-cell RNA-seq applied to a viral human cancer: applications to HPV expression and splicing analysis in HeLa S3 cells
- PMID: 26550473
- PMCID: PMC4635585
- DOI: 10.1186/s13742-015-0091-4
Full-length single-cell RNA-seq applied to a viral human cancer: applications to HPV expression and splicing analysis in HeLa S3 cells
Abstract
Background: Viral infection causes multiple forms of human cancer, and HPV infection is the primary factor in cervical carcinomas. Recent single-cell RNA-seq studies highlight the tumor heterogeneity present in most cancers, but virally induced tumors have not been studied. HeLa is a well characterized HPV+ cervical cancer cell line.
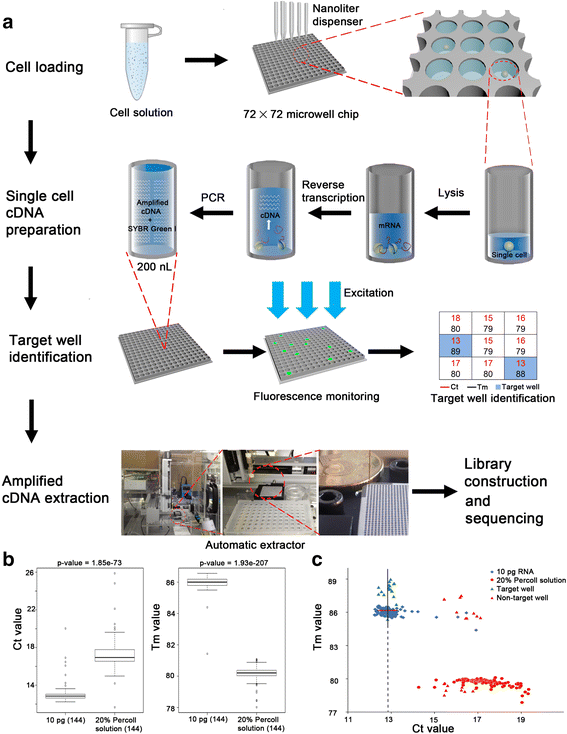
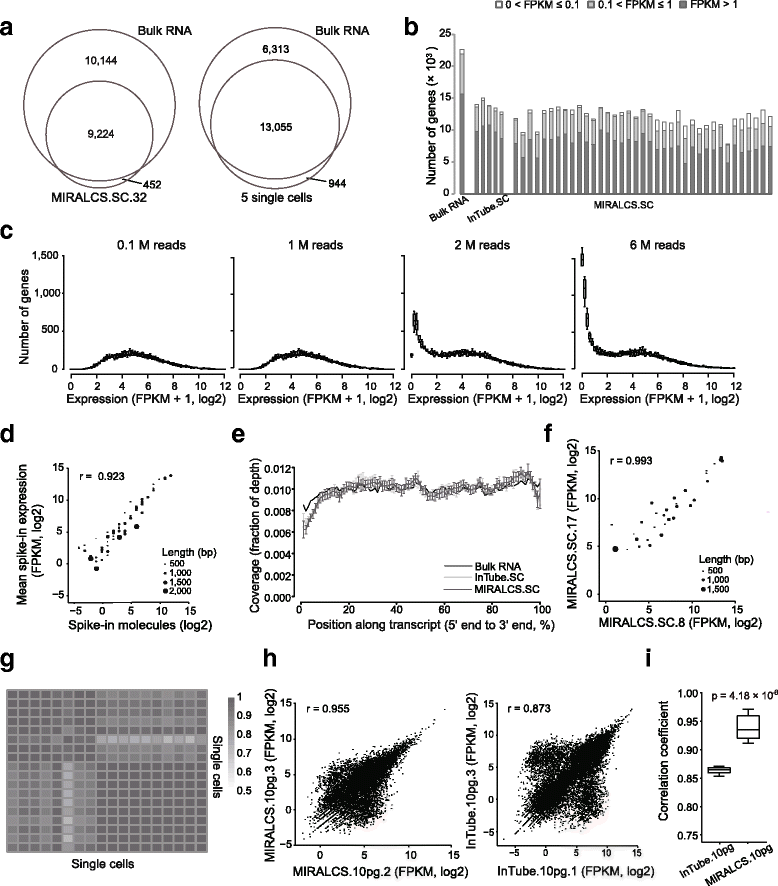
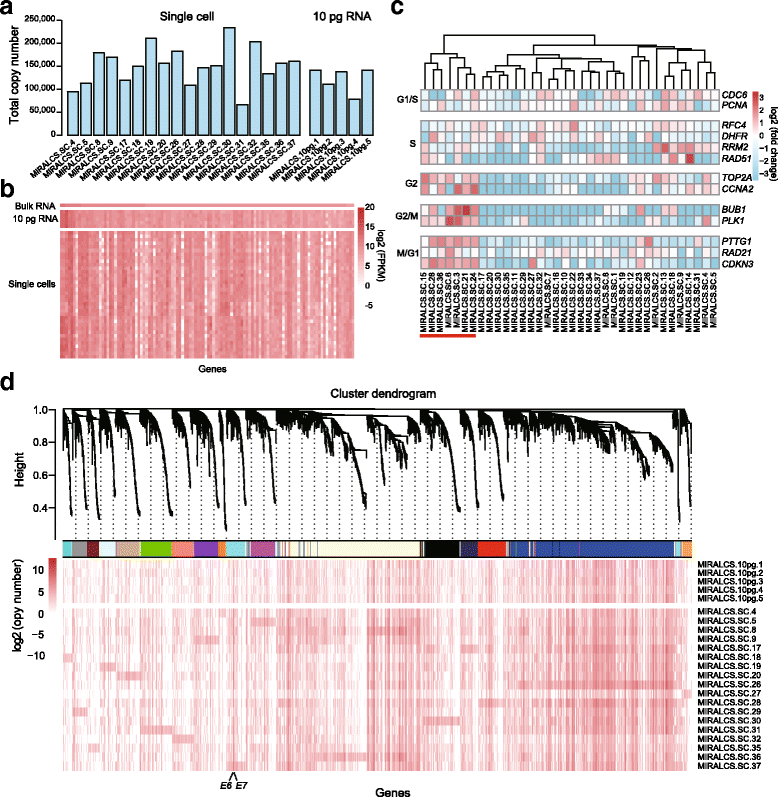
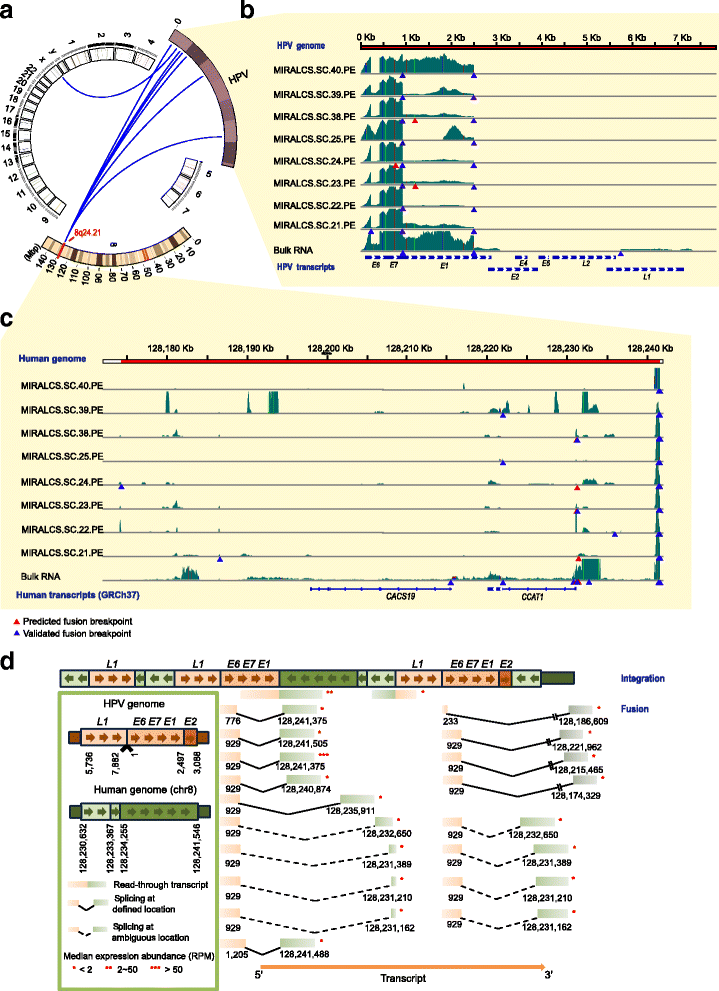
Result: We developed a new high throughput platform to prepare single-cell RNA on a nanoliter scale based on a customized microwell chip. Using this method, we successfully amplified full-length transcripts of 669 single HeLa S3 cells and 40 of them were randomly selected to perform single-cell RNA sequencing. Based on these data, we obtained a comprehensive understanding of the heterogeneity of HeLa S3 cells in gene expression, alternative splicing and fusions. Furthermore, we identified a high diversity of HPV-18 expression and splicing at the single-cell level. By co-expression analysis we identified 283 E6, E7 co-regulated genes, including CDC25, PCNA, PLK4, BUB1B and IRF1 known to interact with HPV viral proteins.
Conclusion: Our results reveal the heterogeneity of a virus-infected cell line. It not only provides a transcriptome characterization of HeLa S3 cells at the single cell level, but is a demonstration of the power of single cell RNA-seq analysis of virally infected cells and cancers.
Keywords: Cancer; HPV; HeLa; RNA splicing; Single-cell transcriptome; Tumor heterogeneity; Virus.
Figures

References
-
- Boyle P, Levin B. World cancer report 2008. Lyon: International Agency for Research on Cancer and World Health Organization Press; 2008.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

