Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes
- PMID: 26551670
- PMCID: PMC4829974
- DOI: 10.1038/ng.3444
Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes
Abstract
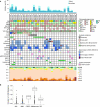
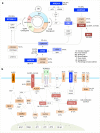
Sézary syndrome is a rare leukemic form of cutaneous T cell lymphoma characterized by generalized redness, scaling, itching and increased numbers of circulating atypical T lymphocytes. It is rarely curable, with poor prognosis. Here we present a multiplatform genomic analysis of 37 patients with Sézary syndrome that implicates dysregulation of cell cycle checkpoint and T cell signaling. Frequent somatic alterations were identified in TP53, CARD11, CCR4, PLCG1, CDKN2A, ARID1A, RPS6KA1 and ZEB1. Activating CCR4 and CARD11 mutations were detected in nearly one-third of patients. ZEB1, encoding a transcription repressor essential for T cell differentiation, was deleted in over one-half of patients. IL32 and IL2RG were overexpressed in nearly all cases. Our results demonstrate profound disruption of key signaling pathways in Sézary syndrome and suggest potential targets for new therapies.
Figures




References
-
- Girardi M, Heald PW, Wilson LD. The pathogenesis of mycosis fungoides. N Engl J Med. 2004;350:1978–88. - PubMed
-
- Kim YH, Liu HL, Mraz-Gernhard S, Varghese A, Hoppe RT. Long-term outcome of 525 patients with mycosis fungoides and Sezary syndrome: clinical prognostic factors and risk for disease progression. Arch Dermatol. 2003;139:857–66. - PubMed
-
- Olsen E, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood. 2007;110:1713–22. - PubMed
-
- Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sézary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. Journal of the American Academy of Dermatology. 2014;70:205, e1–16. quiz 221-222. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

