Functions of TET Proteins in Hematopoietic Transformation
- PMID: 26552488
- PMCID: PMC4673406
- DOI: 10.14348/molcells.2015.0294
Functions of TET Proteins in Hematopoietic Transformation
Abstract
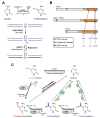
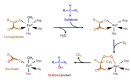
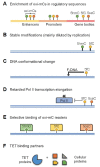
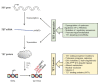
DNA methylation is a well-characterized epigenetic modification that plays central roles in mammalian development, genomic imprinting, X-chromosome inactivation and silencing of retrotransposon elements. Aberrant DNA methylation pattern is a characteristic feature of cancers and associated with abnormal expression of oncogenes, tumor suppressor genes or repair genes. Ten-eleven-translocation (TET) proteins are recently characterized dioxygenases that catalyze progressive oxidation of 5-methylcytosine to produce 5-hydroxymethylcytosine and further oxidized derivatives. These oxidized methylcytosines not only potentiate DNA demethylation but also behave as independent epigenetic modifications per se. The expression or activity of TET proteins and DNA hydroxymethylation are highly dysregulated in a wide range of cancers including hematologic and non-hematologic malignancies, and accumulating evidence points TET proteins as a novel tumor suppressor in cancers. Here we review DNA demethylation-dependent and -independent functions of TET proteins. We also describe diverse TET loss-of-function mutations that are recurrently found in myeloid and lymphoid malignancies and their potential roles in hematopoietic transformation. We discuss consequences of the deficiency of individual Tet genes and potential compensation between different Tet members in mice. Possible mechanisms underlying facilitated oncogenic transformation of TET-deficient hematopoietic cells are also described. Lastly, we address non-mutational mechanisms that lead to suppression or inactivation of TET proteins in cancers. Strategies to restore normal 5mC oxidation status in cancers by targeting TET proteins may provide new avenues to expedite the development of promising anti-cancer agents.
Keywords: 5-methylcytosine oxidation; TET protein; hematologic malignancies; hematopoiesis; tumor suppression.
Figures
Similar articles
-
TET proteins and 5-methylcytosine oxidation in hematological cancers.Immunol Rev. 2015 Jan;263(1):6-21. doi: 10.1111/imr.12239. Immunol Rev. 2015. PMID: 25510268 Free PMC article. Review.
-
Dysregulation of the TET family of epigenetic regulators in lymphoid and myeloid malignancies.Blood. 2019 Oct 31;134(18):1487-1497. doi: 10.1182/blood.2019791475. Blood. 2019. PMID: 31467060 Free PMC article. Review.
-
Epigenetic Function of TET Family, 5-Methylcytosine, and 5-Hydroxymethylcytosine in Hematologic Malignancies.Oncol Res Treat. 2019;42(6):309-318. doi: 10.1159/000498947. Epub 2019 May 3. Oncol Res Treat. 2019. PMID: 31055566 Review.
-
Epigenetic Modification of Cytosines in Hematopoietic Differentiation and Malignant Transformation.Int J Mol Sci. 2023 Jan 15;24(2):1727. doi: 10.3390/ijms24021727. Int J Mol Sci. 2023. PMID: 36675240 Free PMC article. Review.
-
TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling.Nature. 2016 Oct 27;538(7626):528-532. doi: 10.1038/nature20095. Epub 2016 Oct 19. Nature. 2016. PMID: 27760115
Cited by
-
Inhibition of DNA methyltransferase leads to increased genomic 5-hydroxymethylcytosine levels in hematopoietic cells.FEBS Open Bio. 2018 Feb 23;8(4):584-592. doi: 10.1002/2211-5463.12392. eCollection 2018 Apr. FEBS Open Bio. 2018. PMID: 29632811 Free PMC article.
-
Tumor necrosis factor-α decreases EC-SOD expression through DNA methylation.J Clin Biochem Nutr. 2017 May;60(3):169-175. doi: 10.3164/jcbn.16-111. Epub 2017 Apr 7. J Clin Biochem Nutr. 2017. PMID: 28584398 Free PMC article.
-
TET family dioxygenases and DNA demethylation in stem cells and cancers.Exp Mol Med. 2017 Apr 28;49(4):e323. doi: 10.1038/emm.2017.5. Exp Mol Med. 2017. PMID: 28450733 Free PMC article. Review.
-
TET3 is a positive regulator of mitochondrial respiration in Neuro2A cells.PLoS One. 2024 Jan 16;19(1):e0294187. doi: 10.1371/journal.pone.0294187. eCollection 2024. PLoS One. 2024. PMID: 38227585 Free PMC article.
-
Lower Levels of TET2 Gene Expression, with a Higher Level of TET2 Promoter Methylation in Patients with AML; Evidence for the Role of Aberrant Methylation in AML Pathogenesis.Indian J Hematol Blood Transfus. 2024 Jan;40(1):52-60. doi: 10.1007/s12288-023-01673-y. Epub 2023 Jun 5. Indian J Hematol Blood Transfus. 2024. PMID: 38312186 Free PMC article.
References
-
- Abbas S., Lugthart S., Kavelaars F.G., Schelen A., Koenders J.E., Zeilemaker A., van Putten W.J., Rijneveld A.W., Lowenberg B., Valk P.J. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116:2122–2126. - PubMed
-
- Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. - PubMed
-
- Avvakumov G.V., Walker J.R., Xue S., Li Y.J., Duan S.L., Bronner C., Arrowsmith C.H., Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

