Micro-proteomics with iterative data analysis: Proteome analysis in C. elegans at the single worm level
- PMID: 26552604
- PMCID: PMC4819713
- DOI: 10.1002/pmic.201500264
Micro-proteomics with iterative data analysis: Proteome analysis in C. elegans at the single worm level
Abstract
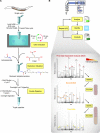
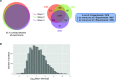
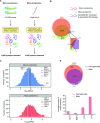
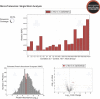
Proteomics studies typically analyze proteins at a population level, using extracts prepared from tens of thousands to millions of cells. The resulting measurements correspond to average values across the cell population and can mask considerable variation in protein expression and function between individual cells or organisms. Here, we report the development of micro-proteomics for the analysis of Caenorhabditis elegans, a eukaryote composed of 959 somatic cells and ∼1500 germ cells, measuring the worm proteome at a single organism level to a depth of ∼3000 proteins. This includes detection of proteins across a wide dynamic range of expression levels (>6 orders of magnitude), including many chromatin-associated factors involved in chromosome structure and gene regulation. We apply the micro-proteomics workflow to measure the global proteome response to heat-shock in individual nematodes. This shows variation between individual animals in the magnitude of proteome response following heat-shock, including variable induction of heat-shock proteins. The micro-proteomics pipeline thus facilitates the investigation of stochastic variation in protein expression between individuals within an isogenic population of C. elegans. All data described in this study are available online via the Encyclopedia of Proteome Dynamics (http://www.peptracker.com/epd), an open access, searchable database resource.
Keywords: Caenorhabditis elegans; Heat-shock; Micro-proteomics; Nematode; Single worm proteomics; Technology.
© 2015 The Authors. PROTEOMICS Published by Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

References
-
- Beck, M. , Claassen, M. , Aebersold, R. , Comprehensive proteomics. Curr. Opin. Biotechnol. 2011, 22, 3–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

