Reconstruction of 3D genome architecture via a two-stage algorithm
- PMID: 26553003
- PMCID: PMC4638111
- DOI: 10.1186/s12859-015-0799-2
Reconstruction of 3D genome architecture via a two-stage algorithm
Abstract
Background: The three-dimensional (3D) configuration of chromosomes within the eukaryote nucleus is an important factor for several cellular functions, including gene expression regulation, and has also been linked with cancer-causing translocation events. While visualization of such architecture remains limited to low resolutions, the ability to infer structures at increasing resolutions has been enabled by recently-devised chromosome conformation capture techniques. In particular, when coupled with next generation sequencing, such methods yield an inventory of genome-wide chromatin contacts or interactions. Various algorithms have been advanced to operate on such contact data to produce reconstructed 3D configurations. Studies have shown that these reconstructions can provide added value over raw interaction data with respect to downstream biological insights. However, only limited, low-resolution reconstructions have been realized for mammals due to computational bottlenecks.
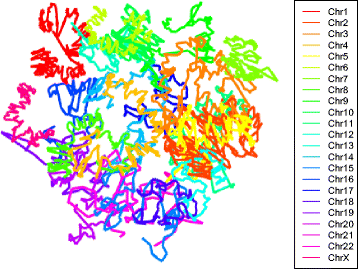
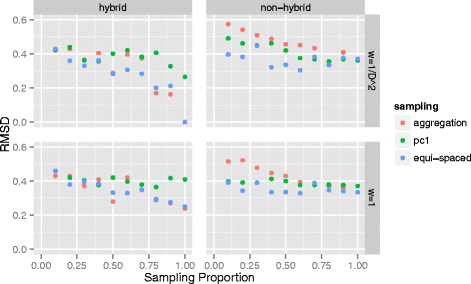
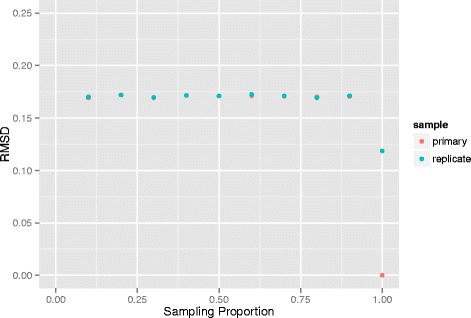
Results: Here we propose a two-stage algorithm to partially overcome these computational barriers. The central idea is to initially utilize existing reconstruction techniques on an individual chromosome basis, using intra-chromosomal contacts, and then to relatively position these chromosome-level reconstructions using inter-chromosomal contacts. This two-stage strategy represents a natural approach in view of the within- versus between- chromosome distribution of contacts. It can increase resolution ≈ 20 fold for mouse and human. After describing the algorithm we present 3D architectures for mouse embryonic stem cells and human lymphoblastoid cells. We evaluate the impact of several factors on reconstruction reproducibility and explore a variety of sampling schemes. We further analyze replicate data at differing resolutions obtained from recently devised in situ Hi-C assays. In all instances we demonstrate insensitivity of the whole-genome 3D reconstruction obtained by the two-stage algorithm to the sampling strategy used.
Conclusions: Our two-stage algorithm has the potential to significantly increase the resolution of 3D genome reconstructions. The improvements are such that we can progress from 1 Mb resolution to 100 kb resolution, notable since this latter value has been identified as critical to inferring topological domains in analyses performed on the contact (rather than 3D) level.
Figures


Similar articles
-
Improved accuracy assessment for 3D genome reconstructions.BMC Bioinformatics. 2018 May 30;19(1):196. doi: 10.1186/s12859-018-2214-2. BMC Bioinformatics. 2018. PMID: 29848293 Free PMC article.
-
Assessing stationary distributions derived from chromatin contact maps.BMC Bioinformatics. 2020 Feb 24;21(1):73. doi: 10.1186/s12859-020-3424-y. BMC Bioinformatics. 2020. PMID: 32093610 Free PMC article.
-
A maximum likelihood algorithm for reconstructing 3D structures of human chromosomes from chromosomal contact data.BMC Genomics. 2018 Feb 23;19(1):161. doi: 10.1186/s12864-018-4546-8. BMC Genomics. 2018. PMID: 29471801 Free PMC article.
-
Computational approaches for inferring 3D conformations of chromatin from chromosome conformation capture data.Methods. 2020 Oct 1;181-182:24-34. doi: 10.1016/j.ymeth.2019.08.008. Epub 2019 Aug 27. Methods. 2020. PMID: 31470090 Free PMC article. Review.
-
A (3D-Nuclear) Space Odyssey: Making Sense of Hi-C Maps.Genes (Basel). 2019 May 29;10(6):415. doi: 10.3390/genes10060415. Genes (Basel). 2019. PMID: 31146487 Free PMC article. Review.
Cited by
-
GenomeFlow: a comprehensive graphical tool for modeling and analyzing 3D genome structure.Bioinformatics. 2019 Apr 15;35(8):1416-1418. doi: 10.1093/bioinformatics/bty802. Bioinformatics. 2019. PMID: 30215673 Free PMC article.
-
Subnuclear distribution of proteins: Links with genome architecture.Nucleus. 2018 Jan 1;9(1):42-55. doi: 10.1080/19491034.2017.1361578. Epub 2017 Sep 14. Nucleus. 2018. PMID: 28910577 Free PMC article. Review.
-
Network analysis identifies chromosome intermingling regions as regulatory hotspots for transcription.Proc Natl Acad Sci U S A. 2017 Dec 26;114(52):13714-13719. doi: 10.1073/pnas.1708028115. Epub 2017 Dec 11. Proc Natl Acad Sci U S A. 2017. PMID: 29229825 Free PMC article.
-
Can 3D diploid genome reconstruction from unphased Hi-C data be salvaged?NAR Genom Bioinform. 2022 May 12;4(2):lqac038. doi: 10.1093/nargab/lqac038. eCollection 2022 Jun. NAR Genom Bioinform. 2022. PMID: 35571676 Free PMC article.
-
Does multi-way, long-range chromatin contact data advance 3D genome reconstruction?BMC Bioinformatics. 2023 Feb 24;24(1):64. doi: 10.1186/s12859-023-05170-x. BMC Bioinformatics. 2023. PMID: 36829114 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

