The Bacterial Enzyme IdeS Cleaves the IgG-Type of B Cell Receptor (BCR), Abolishes BCR-Mediated Cell Signaling, and Inhibits Memory B Cell Activation
- PMID: 26553074
- PMCID: PMC4671093
- DOI: 10.4049/jimmunol.1501929
The Bacterial Enzyme IdeS Cleaves the IgG-Type of B Cell Receptor (BCR), Abolishes BCR-Mediated Cell Signaling, and Inhibits Memory B Cell Activation
Abstract
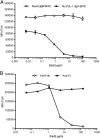
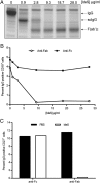
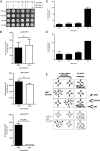
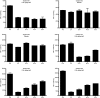
Ag binding to the BCR is a critical step in B cell development and activation, initiating a cascade of signaling events ultimately leading to proliferation, differentiation, or cell death. A bacterial enzyme, IgG-degrading enzyme of Streptococcus pyogenes (IdeS), was shown to specifically cleave IgG molecules below the hinge region of soluble IgG and when IgG is bound to Ag, resulting in one F(ab')2 molecule and one homodimeric Fc fragment. Whether IdeS could also cleave the IgG molecule when it is present in the BCR attached to the B cell membrane in a complex with CD79a and CD79b is unknown. In this article, we present human in vitro and ex vivo data showing that IdeS cleaves the IgG present in the BCR complex and very efficiently blocks Ag binding to the BCR. As a consequence of IdeS cleaving the BCR, signaling cascades downstream of the BCR are blocked, and memory B cells are temporarily silenced, preventing them from responding to antigenic stimulation and their transition into Ab-producing cells.
Copyright © 2015 by The American Association of Immunologists, Inc.
Figures




References
-
- Su Y. F., Chuang W. J., Wang S. M., Chen W. Y., Chiang-Ni C., Lin Y. S., Wu J. J., Liu C. C. 2011. The deficient cleavage of M protein-bound IgG by IdeS: insight into the escape of Streptococcus pyogenes from antibody-mediated immunity. Mol. Immunol. 49: 134–142. - PubMed
-
- Reth M. 1989. Antigen receptor tail clue. Nature 338: 383–384. - PubMed
-
- Reth M., Wienands J. 1997. Initiation and processing of signals from the B cell antigen receptor. Annu. Rev. Immunol. 15: 453–479. - PubMed
-
- Kurosaki T. 2002. Regulation of B-cell signal transduction by adaptor proteins. Nat. Rev. Immunol. 2: 354–363. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

