Construction of a high-density linkage map and fine mapping of QTL for growth in Asian seabass
- PMID: 26553309
- PMCID: PMC4639833
- DOI: 10.1038/srep16358
Construction of a high-density linkage map and fine mapping of QTL for growth in Asian seabass
Abstract
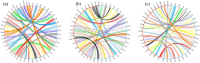
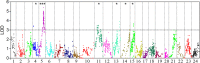
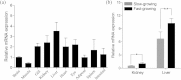
A high-density genetic map is essential for comparative genomic studies and fine mapping of QTL, and can also facilitate genome sequence assembly. Here, a high density genetic map of Asian seabass was constructed with 3321 SNPs generated by sequencing 144 individuals in a F2 family. The length of the map was 1577.67 cM with an average marker interval of 0.52 cM. A high level of genomic synteny among Asian seabass, European seabass, Nile tilapia and stickleback was detected. Using this map, one genome-wide significant and five suggestive QTL for growth traits were detected in six linkage groups (i.e. LG4, LG5, LG11, LG13, LG14 and LG15). These QTL explained 10.5-16.0% of phenotypic variance. A candidate gene, ACOX1 within the significant QTL on LG5 was identified. The gene was differentially expressed between fast- and slow-growing Asian seabass. The high-density SNP-based map provides an important tool for fine mapping QTL in molecular breeding and comparative genome analysis.
Figures

References
-
- Yue G. H. Recent advances of genome mapping and marker‐assisted selection in aquaculture. Fish. 15, 376–396 (2014).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

