3D genome organization in health and disease: emerging opportunities in cancer translational medicine
- PMID: 26553406
- PMCID: PMC4915485
- DOI: 10.1080/19491034.2015.1106676
3D genome organization in health and disease: emerging opportunities in cancer translational medicine
Abstract
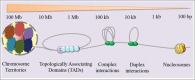
Organizing the DNA to fit inside a spatially constrained nucleus is a challenging problem that has attracted the attention of scientists across all disciplines of science. Increasing evidence has demonstrated the importance of genome geometry in several cellular contexts that affect human health. Among several approaches, the application of sequencing technologies has substantially increased our understanding of this intricate organization, also known as chromatin interactions. These structures are involved in transcriptional control of gene expression by connecting distal regulatory elements with their target genes and regulating co-transcriptional splicing. In addition, chromatin interactions play pivotal roles in the organization of the genome, the formation of structural variants, recombination, DNA replication and cell division. Mutations in factors that regulate chromatin interactions lead to the development of pathological conditions, for example, cancer. In this review, we discuss key findings that have shed light on the importance of these structures in the context of cancers, and highlight the applicability of chromatin interactions as potential biomarkers in molecular medicine as well as therapeutic implications of chromatin interactions.
Keywords: 3D genome organization; ChIA-PET; biomarkers; cancer; chromatin interactions; translational medicine.
Figures


References
-
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol 2010; 2(3):a003889; PMID:20300217; http://dx.doi.org/ 10.1101/cshperspect.a003889 - DOI - PMC - PubMed
-
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al.. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 2009; 41(8):882-4; PMID:19561607; http://dx.doi.org/ 10.1038/ng.403 - DOI - PMC - PubMed
-
- Pittman AM, Naranjo S, Jalava SE, Twiss P, Ma Y, Olver B, Lloyd A, Vijayakrishnan J, Qureshi M, Broderick P, et al.. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet 2010; 6(9):e1001126; PMID:20862326; http://dx.doi.org/ 10.1371/journal.pgen.1001126 - DOI - PMC - PubMed
-
- Meyer KB, Maia AT, O'Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P, Ambs S, Prokunina-Olsson L, Carroll J, Ponder BA. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet 2011; 7(7):e1002165; PMID:21814516; http://dx.doi.org/ 10.1371/journal.pgen.1002165 - DOI - PMC - PubMed
-
- Ahmadiyeh N,Pomerantz MM, Grisanzio C Paula Herman Jia L Almendro V He HH Brown M Liu XS Davis M et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A, 2010; 107(21); 9742-6; http://dx.doi.org/ 10.1073/pnas.0910668107 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
