Rolling Circle Translation of Circular RNA in Living Human Cells
- PMID: 26553571
- PMCID: PMC4639774
- DOI: 10.1038/srep16435
Rolling Circle Translation of Circular RNA in Living Human Cells
Abstract
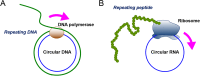
We recently reported that circular RNA is efficiently translated by a rolling circle amplification (RCA) mechanism in a cell-free Escherichia coli translation system. Recent studies have shown that circular RNAs composed of exonic sequences are abundant in human cells. However, whether these circular RNAs can be translated into proteins within cells remains unclear. In this study, we prepared circular RNAs with an infinite open reading frame and tested their translation in eukaryotic systems. Circular RNAs were translated into long proteins in rabbit reticulocyte lysate in the absence of any particular element for internal ribosome entry, a poly-A tail, or a cap structure. The translation systems in eukaryote can accept much simpler RNA as a template for protein synthesis by cyclisation. Here, we demonstrated that the circular RNA is efficiently translated in living human cells to produce abundant protein product by RCA mechanism. These findings suggest that translation of exonic circular RNAs present in human cells is more probable than previously thought.
Figures


References
-
- Abe N. et al.Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew. Chem., Int. Ed. 52, 7004–7008 (2013). - PubMed
-
- Ali M.M. et al.Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 43, 3324–3341 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

