Molecular profiling of 6,892 colorectal cancer samples suggests different possible treatment options specific to metastatic sites
- PMID: 26553611
- PMCID: PMC4847814
- DOI: 10.1080/15384047.2015.1113356
Molecular profiling of 6,892 colorectal cancer samples suggests different possible treatment options specific to metastatic sites
Abstract
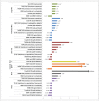
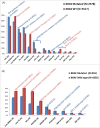
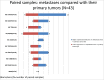
Metastatic colorectal cancer (mCRC) carries a poor prognosis with an overall 5-year survival of 13.1%. Therapies guided by tumor profiling have suggested benefit in advanced cancer. We used a multiplatform molecular profiling (MP) approach to identify key molecular changes that may provide therapeutic options not typically considered in mCRC. We evaluated 6892 mCRC referred to Caris Life Sciences by MP including sequencing (Sanger/NGS), immunohistochemistry (IHC) and in-situ hybridization (ISH). mCRC metastases to liver, brain, ovary or lung (n = 1507) showed differential expression of markers including high protein expression of TOPO1 (52%) and/or low RRM1 (57%), TS (71%) and MGMT (39%), suggesting possible benefit from irinotecan, gemcitabine, 5FU/capecitabine and temozolomide, respectively. Lung metastases harbored a higher Her2 protein expression than the primary colon tumors (4% vs. 1.8%, p = 0.028). Brain and lung metastases had higher KRAS mutations than other sites (65% vs 59% vs 47%, respectively, p = 0.07, <0.01), suggesting poor response to anti-EGFR therapies. BRAF-mutated CRC (n = 455) showed coincident high protein expression of RRM1 (56%), TS (53%) and low PDGFR (22%) as compared with BRAF wild-type tumors. KRAS-mutated mCRC had higher protein expression of c-MET (47% vs. 36%) and lower MGMT (56% vs. 63%), suggesting consideration of c-MET inhibitors and temozolomide. KRAS-mutated CRC had high TUBB3 (42% vs. 33%) and low Her2 by IHC (0.5%) and HER2 by FISH (3%, p <0.05). CRC primaries had a lower incidence of PIK3CA and BRAF mutations in rectal cancer versus colon cancer (10% and 3.3%, respectively). MP of 6892 CRCs identified significant differences between primary and metastatic sites and among BRAF/KRAS sub-types. Our findings are hypothesis generating and need to be examined in prospective studies. Specific therapies may be considered for different actionable targets in mCRC as revealed by MP.
Keywords: APC; BRAF; Cox2; ERCC1; Her2; KRAS; TP53; Topo1; c-MET; colorectal cancer; irinotecan; metastasis; molecular profiling; oxaliplatin; peritoneal cancer; precision medicine.
Figures

References
-
- Jameson GS, Petricoin EF, Sachdev J, Liotta LA, Loesch DM, Anthony SP, Chadha MK, Wulfkuhle JD, Gallagher RI, Reeder KA, et al. . A pilot study utilizing multi-omic molecular profiling to find potential targets and select individualized treatments for patients with previously treated metastatic breast cancer. Breast Cancer Res Treat 2014; 147(3):579–88; PMID:25209003; http://dx.doi.org/10.1007/s10549-014-3117-1 - DOI - PubMed
-
- Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Luthra R, et al. . Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012; 18(22):6373–83; PMID:22966018; http://dx.doi.org/10.1158/1078-0432.CCR-12-1627 - DOI - PMC - PubMed
-
- Tsimberidou AM, Wen S, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Aldape K, et al. . Personalized medicine for patients with advanced cancer in the phase I program at MD anderson: validation and landmark analyses. Clin Cancer Res 2014; 20(18):4827–36; PMID:24987059; http://dx.doi.org/10.1158/1078-0432.CCR-14-0603 - DOI - PMC - PubMed
-
- Von Hoff DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, Korn RL, Desai N, Trieu V, Iglesias JL, et al. . Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol 2011; 29(34):4548–54; PMID:21969517; http://dx.doi.org/10.1200/JCO.2011.36.5742 - DOI - PMC - PubMed
-
- Van Cutsem E, Nowacki M, Lang I, Cascinu S, Shchepotin I, Maurel J, Rougier P, Cunningham D, Nippgen J, Köhneet C. Randomized phase III study of irinotecan and 5-FU/FA with or without cetuximab in the first-line treatment of patients with metastatic colorectal cancer (mCRC): The CRYSTAL trial. J Clin Oncol 2007; 25(18S):4000
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
