Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load
- PMID: 26553974
- PMCID: PMC4664299
- DOI: 10.1073/pnas.1514867112
Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load
Abstract
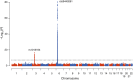
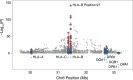
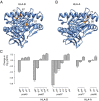
Previous genome-wide association studies (GWAS) of HIV-1-infected populations have been underpowered to detect common variants with moderate impact on disease outcome and have not assessed the phenotypic variance explained by genome-wide additive effects. By combining the majority of available genome-wide genotyping data in HIV-infected populations, we tested for association between ∼8 million variants and viral load (HIV RNA copies per milliliter of plasma) in 6,315 individuals of European ancestry. The strongest signal of association was observed in the HLA class I region that was fully explained by independent effects mapping to five variable amino acid positions in the peptide binding grooves of the HLA-B and HLA-A proteins. We observed a second genome-wide significant association signal in the chemokine (C-C motif) receptor (CCR) gene cluster on chromosome 3. Conditional analysis showed that this signal could not be fully attributed to the known protective CCR5Δ32 allele and the risk P1 haplotype, suggesting further causal variants in this region. Heritability analysis demonstrated that common human genetic variation-mostly in the HLA and CCR5 regions-explains 25% of the variability in viral load. This study suggests that analyses in non-European populations and of variant classes not assessed by GWAS should be priorities for the field going forward.
Keywords: GWAS; HIV-1 control; genomics; heritability; infectious disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mellors JW, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122(8):573–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

