Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives
- PMID: 26553979
- PMCID: PMC4664295
- DOI: 10.1073/pnas.1516925112
Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives
Abstract
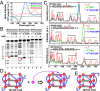
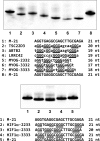
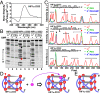
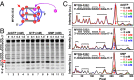
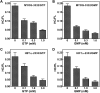
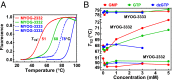
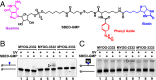
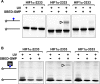
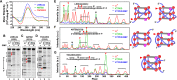
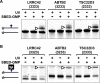
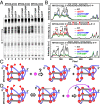
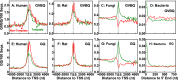
G-quadruplex structures formed by guanine-rich nucleic acids are implicated in essential physiological and pathological processes and nanodevices. G-quadruplexes are normally composed of four Gn (n ≥ 3) tracts assembled into a core of multiple stacked G-quartet layers. By dimethyl sulfate footprinting, circular dichroism spectroscopy, thermal melting, and photo-cross-linking, here we describe a unique type of intramolecular G-quadruplex that forms with one G2 and three G3 tracts and bears a guanine vacancy (G-vacancy) in one of the G-quartet layers. The G-vacancy can be filled up by a guanine base from GTP or GMP to complete an intact G-quartet by Hoogsteen hydrogen bonding, resulting in significant G-quadruplex stabilization that can effectively alter DNA replication in vitro at physiological concentration of GTP and Mg(2+). A bioinformatic survey shows motifs of such G-quadruplexes are evolutionally selected in genes with unique distribution pattern in both eukaryotic and prokaryotic organisms, implying such G-vacancy-bearing G-quadruplexes are present and play a role in gene regulation. Because guanine derivatives are natural metabolites in cells, the formation of such G-quadruplexes and guanine fill-in (G-fill-in) may grant an environment-responsive regulation in cellular processes. Our findings thus not only expand the sequence definition of G-quadruplex formation, but more importantly, reveal a structural and functional property not seen in the standard canonical G-quadruplexes.
Keywords: G-quadruplex; G-vacancy; guanine-responsive; nucleic acids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Tarsounas M, Tijsterman M. Genomes and G-quadruplexes: For better or for worse. J Mol Biol. 2013;425(23):4782–4789. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

