Noninvasive detection of fetal subchromosomal abnormalities by semiconductor sequencing of maternal plasma DNA
- PMID: 26554006
- PMCID: PMC4664371
- DOI: 10.1073/pnas.1518151112
Noninvasive detection of fetal subchromosomal abnormalities by semiconductor sequencing of maternal plasma DNA
Abstract
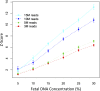
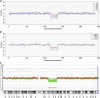
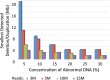
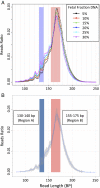
Noninvasive prenatal testing (NIPT) using sequencing of fetal cell-free DNA from maternal plasma has enabled accurate prenatal diagnosis of aneuploidy and become increasingly accepted in clinical practice. We investigated whether NIPT using semiconductor sequencing platform (SSP) could reliably detect subchromosomal deletions/duplications in women carrying high-risk fetuses. We first showed that increasing concentration of abnormal DNA and sequencing depth improved detection. Subsequently, we analyzed plasma from 1,456 pregnant women to develop a method for estimating fetal DNA concentration based on the size distribution of DNA fragments. Finally, we collected plasma from 1,476 pregnant women with fetal structural abnormalities detected on ultrasound who also underwent an invasive diagnostic procedure. We used SSP of maternal plasma DNA to detect subchromosomal abnormalities and validated our results with array comparative genomic hybridization (aCGH). With 3.5 million reads, SSP detected 56 of 78 (71.8%) subchromosomal abnormalities detected by aCGH. With increased sequencing depth up to 10 million reads and restriction of the size of abnormalities to more than 1 Mb, sensitivity improved to 69 of 73 (94.5%). Of 55 false-positive samples, 35 were caused by deletions/duplications present in maternal DNA, indicating the necessity of a validation test to exclude maternal karyotype abnormalities. This study shows that detection of fetal subchromosomal abnormalities is a viable extension of NIPT based on SSP. Although we focused on the application of cell-free DNA sequencing for NIPT, we believe that this method has broader applications for genetic diagnosis, such as analysis of circulating tumor DNA for detection of cancer.
Keywords: NIPT; cell-free DNA; maternal plasma DNA; noninvasive prenatal testing; semiconductor sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
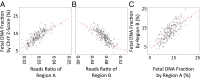
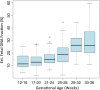
Similar articles
-
Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: implications for cancer screening.BMC Med. 2016 Aug 24;14(1):126. doi: 10.1186/s12916-016-0667-6. BMC Med. 2016. PMID: 27558279 Free PMC article.
-
Noninvasive Prenatal Testing of Rare Autosomal Aneuploidies by Semiconductor Sequencing.DNA Cell Biol. 2018 Mar;37(3):174-181. doi: 10.1089/dna.2017.4075. Epub 2018 Jan 30. DNA Cell Biol. 2018. PMID: 29381401
-
Detection of complex deletions in chromosomes 13 and 21 in a fetus by noninvasive prenatal testing.Mol Cytogenet. 2016 Jan 12;9:3. doi: 10.1186/s13039-016-0213-4. eCollection 2016. Mol Cytogenet. 2016. PMID: 26759606 Free PMC article.
-
Noninvasive prenatal testing using cell-free fetal DNA in maternal plasma.Curr Protoc Hum Genet. 2015 Jan 20;84:8.15.1-8.15.20. doi: 10.1002/0471142905.hg0815s84. Curr Protoc Hum Genet. 2015. PMID: 25599670 Review.
-
Prenatal Diagnosis Innovation: Genome Sequencing of Maternal Plasma.Annu Rev Med. 2016;67:419-32. doi: 10.1146/annurev-med-091014-115715. Epub 2015 Oct 15. Annu Rev Med. 2016. PMID: 26473414 Review.
Cited by
-
The performance evaluation of NIPT for fetal chromosome microdeletion/microduplication detection: a retrospective analysis of 68,588 Chinese cases.Front Genet. 2024 Jun 7;15:1390539. doi: 10.3389/fgene.2024.1390539. eCollection 2024. Front Genet. 2024. PMID: 38911296 Free PMC article.
-
Prenatal genetic detection in foetus with gallbladder size anomalies: cohort study and systematic review of the literature.Ann Med. 2025 Dec;57(1):2440638. doi: 10.1080/07853890.2024.2440638. Epub 2024 Dec 13. Ann Med. 2025. PMID: 39670967 Free PMC article.
-
Efficiency of expanded noninvasive prenatal testing in the detection of fetal subchromosomal microdeletion and microduplication in a cohort of 31,256 single pregnancies.Sci Rep. 2022 Nov 17;12(1):19750. doi: 10.1038/s41598-022-24337-9. Sci Rep. 2022. PMID: 36396840 Free PMC article.
-
Performances of NIPT for copy number variations at different sequencing depths using the semiconductor sequencing platform.Hum Genomics. 2021 Jul 2;15(1):41. doi: 10.1186/s40246-021-00332-5. Hum Genomics. 2021. PMID: 34215332 Free PMC article.
-
Evaluation of non-invasive prenatal testing to detect chromosomal aberrations in a Chinese cohort.J Cell Mol Med. 2019 Nov;23(11):7873-7878. doi: 10.1111/jcmm.14614. Epub 2019 Aug 27. J Cell Mol Med. 2019. PMID: 31454164 Free PMC article.
References
-
- Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8(8):639–646. - PubMed
-
- Lejeune J, et al. 3 Cases of partial deletion of the short arm of a 5 chromosome. C R Hebd Seances Acad Sci. 1963;257:3098–3102. - PubMed
-
- Lammer EJ, Opitz JM. The DiGeorge anomaly as a developmental field defect. Am J Med Genet Suppl. 1986;2:113–127. - PubMed
-
- Lee CN, et al. Clinical utility of array comparative genomic hybridisation for prenatal diagnosis: A cohort study of 3171 pregnancies. BJOG. 2012;119(5):614–625. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

