Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver
- PMID: 26554015
- PMCID: PMC4664316
- DOI: 10.1073/pnas.1515308112
Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver
Abstract
Diurnal oscillations of gene expression are a hallmark of rhythmic physiology across most living organisms. Such oscillations are controlled by the interplay between the circadian clock and feeding rhythms. Although rhythmic mRNA accumulation has been extensively studied, comparatively less is known about their transcription and translation. Here, we quantified simultaneously temporal transcription, accumulation, and translation of mouse liver mRNAs under physiological light-dark conditions and ad libitum or night-restricted feeding in WT and brain and muscle Arnt-like 1 (Bmal1)-deficient animals. We found that rhythmic transcription predominantly drives rhythmic mRNA accumulation and translation for a majority of genes. Comparison of wild-type and Bmal1 KO mice shows that circadian clock and feeding rhythms have broad impact on rhythmic gene expression, Bmal1 deletion affecting surprisingly both transcriptional and posttranscriptional levels. Translation efficiency is differentially regulated during the diurnal cycle for genes with 5'-Terminal Oligo Pyrimidine tract (5'-TOP) sequences and for genes involved in mitochondrial activity, many harboring a Translation Initiator of Short 5'-UTR (TISU) motif. The increased translation efficiency of 5'-TOP and TISU genes is mainly driven by feeding rhythms but Bmal1 deletion also affects amplitude and phase of translation, including TISU genes. Together this study emphasizes the complex interconnections between circadian and feeding rhythms at several steps ultimately determining rhythmic gene expression and translation.
Keywords: 5′-TOP sequences; TISU motifs; circadian rhythms; mRNA translation; ribosome profiling.
Conflict of interest statement
Conflict of interest statement: F.A., C.G., J.M., E.M., B.W., G.L., P.D., and F.G. are employees of Nestlé Institute of Health Sciences SA.
Figures

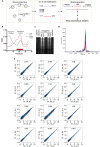
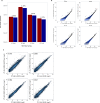
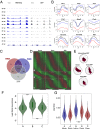
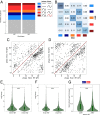




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

