A Naturally Occurring rev1-vpu Fusion Gene Does Not Confer a Fitness Advantage to HIV-1
- PMID: 26554585
- PMCID: PMC4640844
- DOI: 10.1371/journal.pone.0142118
A Naturally Occurring rev1-vpu Fusion Gene Does Not Confer a Fitness Advantage to HIV-1
Abstract
Background: Pandemic strains of HIV-1 (group M) encode a total of nine structural (gag, pol, env), regulatory (rev, tat) and accessory (vif, vpr, vpu, nef) genes. However, some subtype A and C viruses exhibit an unusual gene arrangement in which the first exon of rev (rev1) and the vpu gene are placed in the same open reading frame. Although this rev1-vpu gene fusion is present in a considerable fraction of HIV-1 strains, its functional significance is unknown.
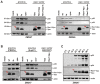
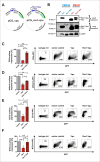
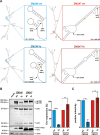
Results: Examining infectious molecular clones (IMCs) of HIV-1 that encode the rev1-vpu polymorphism, we show that a fusion protein is expressed in infected cells. Due to the splicing pattern of viral mRNA, however, these same IMCs also express a regular Vpu protein, which is produced at much higher levels. To investigate the function of the fusion gene, we characterized isogenic IMC pairs differing only in their ability to express a Rev1-Vpu protein. Analysis in transfected HEK293T and infected CD4+ T cells showed that all of these viruses were equally active in known Vpu functions, such as down-modulation of CD4 or counteraction of tetherin. Furthermore, the polymorphism did not affect Vpu-mediated inhibition of NF-кB activation or Rev-dependent nuclear export of incompletely spliced viral mRNAs. There was also no evidence for enhanced replication of Rev1-Vpu expressing viruses in primary PBMCs or ex vivo infected human lymphoid tissues. Finally, the frequency of HIV-1 quasispecies members that encoded a rev1-vpu fusion gene did not change in HIV-1 infected individuals over time.
Conclusions: Expression of a rev1-vpu fusion gene does not affect regular Rev and Vpu functions or alter HIV-1 replication in primary target cells. Since there is no evidence for increased replication fitness of rev1-vpu encoding viruses, this polymorphism likely emerged in the context of other mutations within and/or outside the rev1-vpu intergenic region, and may have a neutral phenotype.
Conflict of interest statement
Figures





Similar articles
-
Unusual Fusion Proteins of HIV-1.Front Microbiol. 2017 Jan 9;7:2152. doi: 10.3389/fmicb.2016.02152. eCollection 2016. Front Microbiol. 2017. PMID: 28119676 Free PMC article. Review.
-
A rev1-vpu polymorphism unique to HIV-1 subtype A and C strains impairs envelope glycoprotein expression from rev-vpu-env cassettes and reduces virion infectivity in pseudotyping assays.Virology. 2010 Feb 20;397(2):346-57. doi: 10.1016/j.virol.2009.11.019. Epub 2009 Dec 8. Virology. 2010. PMID: 20003995 Free PMC article.
-
Viral diversity and diversification of major non-structural genes vif, vpr, vpu, tat exon 1 and rev exon 1 during primary HIV-1 subtype C infection.PLoS One. 2012;7(5):e35491. doi: 10.1371/journal.pone.0035491. Epub 2012 May 9. PLoS One. 2012. PMID: 22590503 Free PMC article. Clinical Trial.
-
Vpu-Mediated Counteraction of Tetherin Is a Major Determinant of HIV-1 Interferon Resistance.mBio. 2016 Aug 16;7(4):e00934-16. doi: 10.1128/mBio.00934-16. mBio. 2016. PMID: 27531907 Free PMC article.
-
Rev-dependent expression of three species of HIV-1 mRNAs (review).Int J Mol Med. 1999 Mar;3(3):297-302. doi: 10.3892/ijmm.3.3.297. Int J Mol Med. 1999. PMID: 10028055 Review.
Cited by
-
Guanylate binding protein 5: Impairing virion infectivity by targeting retroviral envelope glycoproteins.Small GTPases. 2017 Jan 2;8(1):31-37. doi: 10.1080/21541248.2016.1189990. Epub 2016 Jun 8. Small GTPases. 2017. PMID: 27275775 Free PMC article.
-
SIVcol Nef counteracts SERINC5 by promoting its proteasomal degradation but does not efficiently enhance HIV-1 replication in human CD4+ T cells and lymphoid tissue.PLoS Pathog. 2018 Aug 20;14(8):e1007269. doi: 10.1371/journal.ppat.1007269. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30125328 Free PMC article.
-
Unusual Fusion Proteins of HIV-1.Front Microbiol. 2017 Jan 9;7:2152. doi: 10.3389/fmicb.2016.02152. eCollection 2016. Front Microbiol. 2017. PMID: 28119676 Free PMC article. Review.
-
Efficient Vpu-Mediated Tetherin Antagonism by an HIV-1 Group O Strain.J Virol. 2017 Feb 28;91(6):e02177-16. doi: 10.1128/JVI.02177-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077643 Free PMC article.
-
HIV-1 Vpu restricts Fc-mediated effector functions in vivo.Cell Rep. 2022 Nov 8;41(6):111624. doi: 10.1016/j.celrep.2022.111624. Cell Rep. 2022. PMID: 36351384 Free PMC article.
References
-
- Cohen EA, Lu Y, Göttlinger H, Dehni G, Jalinoos Y, Sodroski JG, et al. The T open reading frame of human immunodeficiency virus type 1. J Acquir Immune Defic Syndr. 1990;3: 601–608. - PubMed
-
- Göttlinger HG, Dorfman T, Cohen EA, Haseltine WA. The role of the tnv protein and tnv RNA splicing signals in replication of HIV-1 IIIB isolates. Virology. 1992;189: 618–628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

