Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment
- PMID: 26555032
- PMCID: PMC4806575
- DOI: 10.1038/hdy.2015.94
Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment
Abstract
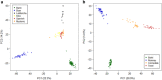
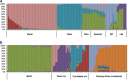
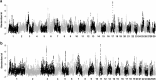
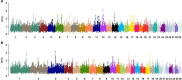
Goats and sheep are versatile domesticates that have been integrated into diverse environments and production systems. Natural and artificial selection have shaped the variation in the two species, but natural selection has played the major role among indigenous flocks. To investigate signals of natural selection, we analyzed genotype data generated using the caprine and ovine 50K SNP BeadChips from Barki goats and sheep that are indigenous to a hot arid environment in Egypt's Coastal Zone of the Western Desert. We identify several candidate regions under selection that spanned 119 genes. A majority of the genes were involved in multiple signaling and signal transduction pathways in a wide variety of cellular and biochemical processes. In particular, selection signatures spanning several genes that directly or indirectly influenced traits for adaptation to hot arid environments, such as thermo-tolerance (melanogenesis) (FGF2, GNAI3, PLCB1), body size and development (BMP2, BMP4, GJA3, GJB2), energy and digestive metabolism (MYH, TRHDE, ALDH1A3), and nervous and autoimmune response (GRIA1, IL2, IL7, IL21, IL1R1) were identified. We also identified eight common candidate genes under selection in the two species and a shared selection signature that spanned a conserved syntenic segment to bovine chromosome 12 on caprine and ovine chromosomes 12 and 10, respectively, providing, most likely, the evidence for selection in a common environment in two different but closely related species. Our study highlights the importance of indigenous livestock as model organisms for investigating selection sweeps and genome-wide association mapping.
Figures
References
-
- Ahtiainen JP, Hulmi JJ, Kraemer WJ, Lohti M, Pakarinen A, Moro AA et al. (2009). Strength, endurance or combined training elicit diverse skeletal muscle myosin heavy chain isoform proportion but unaltered androgen receptor concentration in older men. Int J Sports Med 30: 879–887. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

